The susceptibility of disulfide bonds towards radiation damage may be explained by S⋯O interactions
- PMID: 32939274
- PMCID: PMC7467163
- DOI: 10.1107/S2052252520008520
The susceptibility of disulfide bonds towards radiation damage may be explained by S⋯O interactions
Abstract
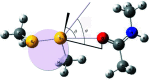
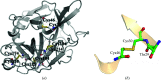
Radiation-induced damage to protein crystals during X-ray diffraction data collection is a major impediment to obtaining accurate structural information on macromolecules. Some of the specific impairments that are inflicted upon highly brilliant X-ray irradiation are metal-ion reduction, disulfide-bond cleavage and a loss of the integrity of the carboxyl groups of acidic residues. With respect to disulfide-bond reduction, previous results have indicated that not all disulfide bridges are equally susceptible to damage. A careful analysis of the chemical environment of disulfide bonds in the structures of elastase, lysozyme, acetylcholinesterase and other proteins suggests that S-S bonds which engage in a close contact with a carbonyl O atom along the extension of the S-S bond vector are more susceptible to reduction than the others. Such an arrangement predisposes electron transfer to occur from the O atom to the disulfide bond, leading to its reduction. The interaction between a nucleophile and an electrophile, akin to hydrogen bonding, stabilizes protein structures, but it also provides a pathway of electron transfer to the S-S bond, leading to its reduction during exposure of the protein crystal to an intense X-ray beam. An otherwise stabilizing interaction can thus be the cause of destabilization under the condition of radiation exposure.
Keywords: NBO; S⋯O interactions; disulfide bonds; electron transfer; quantum-chemical calculations; radiation damage.
© Rajasri Bhattacharyya et al. 2020.
Figures


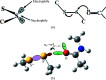
Similar articles
-
Planning Implications Related to Sterilization-Sensitive Science Investigations Associated with Mars Sample Return (MSR).Astrobiology. 2022 Jun;22(S1):S112-S164. doi: 10.1089/AST.2021.0113. Epub 2022 May 19. Astrobiology. 2022. PMID: 34904892
-
The significant role of the intermolecular CH⋯O/N hydrogen bonds in governing the biologically important pairs of the DNA and RNA modified bases: a comprehensive theoretical investigation.J Biomol Struct Dyn. 2015;33(8):1624-52. doi: 10.1080/07391102.2014.968623. Epub 2014 Oct 28. J Biomol Struct Dyn. 2015. PMID: 25350312
-
In Situ Assessment of Intrinsic Strength of X-I⋯OA-Type Halogen Bonds in Molecular Crystals with Periodic Local Vibrational Mode Theory.Molecules. 2020 Mar 30;25(7):1589. doi: 10.3390/molecules25071589. Molecules. 2020. PMID: 32235623 Free PMC article.
-
Evidence for the formation of disulfide radicals in protein crystals upon X-ray irradiation.J Synchrotron Radiat. 2002 Nov 1;9(Pt 6):342-6. doi: 10.1107/s0909049502014589. Epub 2002 Nov 1. J Synchrotron Radiat. 2002. PMID: 12409620
-
The Pnictogen Bond: The Covalently Bound Arsenic Atom in Molecular Entities in Crystals as a Pnictogen Bond Donor.Molecules. 2022 May 25;27(11):3421. doi: 10.3390/molecules27113421. Molecules. 2022. PMID: 35684359 Free PMC article. Review.
Cited by
-
Doses for X-ray and electron diffraction: New features in RADDOSE-3D including intensity decay models.Protein Sci. 2024 Jul;33(7):e5005. doi: 10.1002/pro.5005. Protein Sci. 2024. PMID: 38923423 Free PMC article.
-
Quantum Mechanics/Molecular Mechanics Simulations Distinguish Insulin-Regulated Aminopeptidase Substrate (Oxytocin) and Inhibitor (Angiotensin IV) and Reveal Determinants of Activity and Inhibition.J Chem Inf Model. 2025 Jun 23;65(12):6261-6272. doi: 10.1021/acs.jcim.5c00869. Epub 2025 Jun 11. J Chem Inf Model. 2025. PMID: 40495656 Free PMC article.
-
Identifying and avoiding radiation damage in macromolecular crystallography.Acta Crystallogr D Struct Biol. 2024 May 1;80(Pt 5):314-327. doi: 10.1107/S2059798324003243. Epub 2024 Apr 30. Acta Crystallogr D Struct Biol. 2024. PMID: 38700059 Free PMC article.
-
Chaperone-mediated MHC-I peptide exchange in antigen presentation.IUCrJ. 2024 May 1;11(Pt 3):287-298. doi: 10.1107/S2052252524002768. IUCrJ. 2024. PMID: 38656309 Free PMC article. Review.
-
Conformational coupling of the sialic acid TRAP transporter HiSiaQM with its substrate binding protein HiSiaP.Nat Commun. 2024 Jan 8;15(1):217. doi: 10.1038/s41467-023-44327-3. Nat Commun. 2024. PMID: 38191530 Free PMC article.
References
-
- Bhattacharyya, R., Pal, D. & Chakrabarti, P. (2004). Protein Eng. Des. Sel. 17, 795–808. - PubMed
-
- Bürgi, H.-B., Dunitz, J. D. & Shefter, E. (1973). J. Am. Chem. Soc. 95, 5065–5067.
-
- Burmeister, W. P. (2000). Acta Cryst. D56, 328–341. - PubMed
-
- Carpentier, P., Royant, A., Weik, M. & Bourgeois, D. (2010). Structure, 18, 1410–1419. - PubMed
LinkOut - more resources
Full Text Sources

