Deep Learning in Proteomics
- PMID: 32939979
- PMCID: PMC7757195
- DOI: 10.1002/pmic.201900335
Deep Learning in Proteomics
Abstract
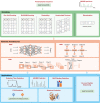
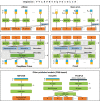
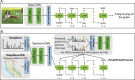
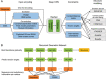
Proteomics, the study of all the proteins in biological systems, is becoming a data-rich science. Protein sequences and structures are comprehensively catalogued in online databases. With recent advancements in tandem mass spectrometry (MS) technology, protein expression and post-translational modifications (PTMs) can be studied in a variety of biological systems at the global scale. Sophisticated computational algorithms are needed to translate the vast amount of data into novel biological insights. Deep learning automatically extracts data representations at high levels of abstraction from data, and it thrives in data-rich scientific research domains. Here, a comprehensive overview of deep learning applications in proteomics, including retention time prediction, MS/MS spectrum prediction, de novo peptide sequencing, PTM prediction, major histocompatibility complex-peptide binding prediction, and protein structure prediction, is provided. Limitations and the future directions of deep learning in proteomics are also discussed. This review will provide readers an overview of deep learning and how it can be used to analyze proteomics data.
Keywords: bioinformatics; deep learning; proteomics.
© 2020 The Authors. Proteomics published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Deep Learning-Based Advances In Protein Posttranslational Modification Site and Protein Cleavage Prediction.Methods Mol Biol. 2022;2499:285-322. doi: 10.1007/978-1-0716-2317-6_15. Methods Mol Biol. 2022. PMID: 35696087
-
π-PrimeNovo: an accurate and efficient non-autoregressive deep learning model for de novo peptide sequencing.Nat Commun. 2025 Jan 2;16(1):267. doi: 10.1038/s41467-024-55021-3. Nat Commun. 2025. PMID: 39747823 Free PMC article.
-
Deep learning approaches for data-independent acquisition proteomics.Expert Rev Proteomics. 2021 Dec;18(12):1031-1043. doi: 10.1080/14789450.2021.2020654. Epub 2021 Dec 28. Expert Rev Proteomics. 2021. PMID: 34918987 Review.
-
Data-independent acquisition proteomics methods for analyzing post-translational modifications.Proteomics. 2023 Apr;23(7-8):e2200046. doi: 10.1002/pmic.202200046. Epub 2022 Sep 7. Proteomics. 2023. PMID: 36036492 Review.
-
Sitetack: a deep learning model that improves PTM prediction by using known PTMs.Bioinformatics. 2024 Nov 1;40(11):btae602. doi: 10.1093/bioinformatics/btae602. Bioinformatics. 2024. PMID: 39388212 Free PMC article.
Cited by
-
Variability analysis of LC-MS experimental factors and their impact on machine learning.Gigascience. 2022 Dec 28;12:giad096. doi: 10.1093/gigascience/giad096. Epub 2023 Nov 20. Gigascience. 2022. PMID: 37983748 Free PMC article.
-
OPDAylation of Thiols of the Redox Regulatory Network In Vitro.Antioxidants (Basel). 2022 Apr 27;11(5):855. doi: 10.3390/antiox11050855. Antioxidants (Basel). 2022. PMID: 35624719 Free PMC article.
-
Managing of Unassigned Mass Spectrometric Data by Neural Network for Cancer Phenotypes Classification.J Pers Med. 2021 Dec 3;11(12):1288. doi: 10.3390/jpm11121288. J Pers Med. 2021. PMID: 34945760 Free PMC article.
-
A tale of solving two computational challenges in protein science: neoantigen prediction and protein structure prediction.Brief Bioinform. 2022 Jan 17;23(1):bbab493. doi: 10.1093/bib/bbab493. Brief Bioinform. 2022. PMID: 34891158 Free PMC article. Review.
-
Proteomics of Penicillium chrysogenum for a Deeper Understanding of Lead (Pb) Metal Bioremediation.ACS Omega. 2024 Jun 6;9(24):26245-26256. doi: 10.1021/acsomega.4c02006. eCollection 2024 Jun 18. ACS Omega. 2024. PMID: 38911750 Free PMC article.
References
-
- Kelchtermans P., Bittremieux W., De Grave K., Degroeve S., Ramon J., Laukens K., Valkenborg D., Barsnes H., Martens L., Proteomics 2014, 14, 353. - PubMed
-
- Bouwmeester R., Gabriels R., Van Den Bossche T., Martens L., Degroeve S., Proteomics 2020, e1900351. - PubMed
-
- Xu L. L., Young A., Zhou A., Rost H. L., Proteomics 2020, e1900352. - PubMed
-
- Ching T., Himmelstein D. S., Beaulieu‐Jones B. K., Kalinin A. A., Do B. T., Way G. P., Ferrero E., Agapow P.‐M., Zietz M., Hoffman M. M., Xie W., Rosen G. L., Lengerich B. J., Israeli J., Lanchantin J., Woloszynek S., Carpenter A. E., Shrikumar A., Xu J., Cofer E. M., Lavender C. A., Turaga S. C., Alexandari A. M., Lu Z., Harris D. J., DeCaprio D., Qi Y., Kundaje A., Peng Y., Wiley L. K., Segler M. H. S., Boca S. M., Swamidass S. J., Huang A., Gitter A., Greene C. S., J. R. Soc. Interface. 2018, 15, 20170387. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

