Scanning window analysis of non-coding regions within normal-tumor whole-genome sequence samples
- PMID: 32940334
- PMCID: PMC8138877
- DOI: 10.1093/bib/bbaa203
Scanning window analysis of non-coding regions within normal-tumor whole-genome sequence samples
Abstract
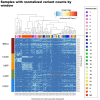
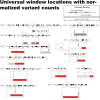
Genomics has benefited from an explosion in affordable high-throughput technology for whole-genome sequencing. The regulatory and functional aspects in non-coding regions may be an important contributor to oncogenesis. Whole-genome tumor-normal paired alignments were used to examine the non-coding regions in five cancer types and two races. Both a sliding window and a binning strategy were introduced to uncover areas of higher than expected variation for additional study. We show that the majority of cancer associated mutations in 154 whole-genome sequences covering breast invasive carcinoma, colon adenocarcinoma, kidney renal papillary cell carcinoma, lung adenocarcinoma and uterine corpus endometrial carcinoma cancers and two races are found outside of the coding region (4 432 885 in non-gene regions versus 1 412 731 in gene regions). A pan-cancer analysis found significantly mutated windows (292 to 3881 in count) demonstrating that there are significant numbers of large mutated regions in the non-coding genome. The 59 significantly mutated windows were found in all studied races and cancers. These offer 16 regions ripe for additional study within 12 different chromosomes-2, 4, 5, 7, 10, 11, 16, 18, 20, 21 and X. Many of these regions were found in centromeric locations. The X chromosome had the largest set of universal windows that cluster almost exclusively in Xq11.1-an area linked to chromosomal instability and oncogenesis. Large consecutive clusters (super windows) were found (19 to 114 in count) providing further evidence that large mutated regions in the genome are influencing cancer development. We show remarkable similarity in highly mutated non-coding regions across both cancer and race.
Keywords: cancer; cancer hotspots; non-coding region; pan-cancer analysis; whole-genome sequencing.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



Similar articles
-
Identification of coding and non-coding mutational hotspots in cancer genomes.BMC Genomics. 2017 Jan 5;18(1):17. doi: 10.1186/s12864-016-3420-9. BMC Genomics. 2017. PMID: 28056774 Free PMC article.
-
Human copy number variants are enriched in regions of low mappability.Nucleic Acids Res. 2018 Aug 21;46(14):7236-7249. doi: 10.1093/nar/gky538. Nucleic Acids Res. 2018. PMID: 30137632 Free PMC article.
-
The FABRIC Cancer Portal: A Ranked Catalogue of Gene Selection in Tumors Over the Human Coding Genome.Cancer Res. 2021 Feb 15;81(4):1178-1185. doi: 10.1158/0008-5472.CAN-20-3147. Epub 2020 Dec 4. Cancer Res. 2021. PMID: 33277365
-
Beyond the coding genome: non-coding mutations and cancer.Front Biosci (Landmark Ed). 2020 Jun 1;25(10):1828-1838. doi: 10.2741/4879. Front Biosci (Landmark Ed). 2020. PMID: 32472759 Free PMC article. Review.
-
Exploring the function of genetic variants in the non-coding genomic regions: approaches for identifying human regulatory variants affecting gene expression.Brief Bioinform. 2015 May;16(3):393-412. doi: 10.1093/bib/bbu018. Epub 2014 Jun 10. Brief Bioinform. 2015. PMID: 24916300 Review.
Cited by
-
Genetic variation and molecular profiling of congenital malformations of the female genital tract based on whole-genome sequencing.World J Pediatr. 2024 Nov;20(11):1179-1195. doi: 10.1007/s12519-024-00839-6. Epub 2024 Sep 9. World J Pediatr. 2024. PMID: 39251565
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

