Quantitative mapping of transcriptome and proteome dynamics during polarization of human iPSC-derived neurons
- PMID: 32940601
- PMCID: PMC7498259
- DOI: 10.7554/eLife.58124
Quantitative mapping of transcriptome and proteome dynamics during polarization of human iPSC-derived neurons
Abstract
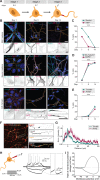
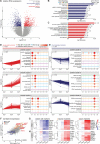
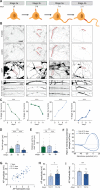
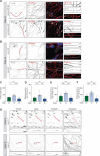
The differentiation of neuronal stem cells into polarized neurons is a well-coordinated process which has mostly been studied in classical non-human model systems, but to what extent these findings are recapitulated in human neurons remains unclear. To study neuronal polarization in human neurons, we cultured hiPSC-derived neurons, characterized early developmental stages, measured electrophysiological responses, and systematically profiled transcriptomic and proteomic dynamics during these steps. The neuron transcriptome and proteome shows extensive remodeling, with differential expression profiles of ~1100 transcripts and ~2200 proteins during neuronal differentiation and polarization. We also identified a distinct axon developmental stage marked by the relocation of axon initial segment proteins and increased microtubule remodeling from the distal (stage 3a) to the proximal (stage 3b) axon. This developmental transition coincides with action potential maturation. Our comprehensive characterization and quantitative map of transcriptome and proteome dynamics provides a solid framework for studying polarization in human neurons.
Keywords: axon; cell biology; development; hiPSC-derived neuron; neuroscience; polarity; proteome; transcriptome.
© 2020, Lindhout et al.
Conflict of interest statement
FL, RK, SP, LH, RS, BS, AA, HM, CW No competing interests declared, CH Employee of Genentech, Inc, a member of the Roche group
Figures





References
-
- Bardy C, van den Hurk M, Kakaradov B, Erwin JA, Jaeger BN, Hernandez RV, Eames T, Paucar AA, Gorris M, Marchand C, Jappelli R, Barron J, Bryant AK, Kellogg M, Lasken RS, Rutten BP, Steinbusch HW, Yeo GW, Gage FH. Predicting the functional states of human iPSC-derived neurons with single-cell RNA-seq and electrophysiology. Molecular Psychiatry. 2016;21:1573–1588. doi: 10.1038/mp.2016.158. - DOI - PMC - PubMed
-
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Research. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. - DOI - PMC - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B. 1995;57:289–300. doi: 10.2307/2346101. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

