Antibody escape by polyomavirus capsid mutation facilitates neurovirulence
- PMID: 32940605
- PMCID: PMC7541085
- DOI: 10.7554/eLife.61056
Antibody escape by polyomavirus capsid mutation facilitates neurovirulence
Abstract
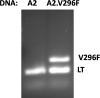
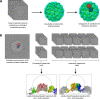
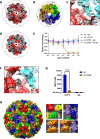
JCPyV polyomavirus, a member of the human virome, causes progressive multifocal leukoencephalopathy (PML), an oft-fatal demyelinating brain disease in individuals receiving immunomodulatory therapies. Mutations in the major viral capsid protein, VP1, are common in JCPyV from PML patients (JCPyV-PML) but whether they confer neurovirulence or escape from virus-neutralizing antibody (nAb) in vivo is unknown. A mouse polyomavirus (MuPyV) with a sequence-equivalent JCPyV-PML VP1 mutation replicated poorly in the kidney, a major reservoir for JCPyV persistence, but retained the CNS infectivity, cell tropism, and neuropathology of the parental virus. This mutation rendered MuPyV resistant to a monoclonal Ab (mAb), whose specificity overlapped the endogenous anti-VP1 response. Using cryo-EM and a custom sub-particle refinement approach, we resolved an MuPyV:Fab complex map to 3.2 Å resolution. The structure revealed the mechanism of mAb evasion. Our findings demonstrate convergence between nAb evasion and CNS neurovirulence in vivo by a frequent JCPyV-PML VP1 mutation.
Keywords: Cryo EM; fab:capsid complex; immunology; inflammation; mouse; neutralizing antibody; polyomavirus; progressive multifocal leukoencephalopathy; subvolume refinement.
© 2020, Lauver et al.
Conflict of interest statement
ML, DG, CN, KA, GJ, DH, EF, SC, CB, SB, NC, SH, AL No competing interests declared
Figures












Similar articles
-
Understanding polyomavirus CNS disease - a perspective from mouse models.FEBS J. 2022 Oct;289(19):5744-5761. doi: 10.1111/febs.16083. Epub 2021 Jul 2. FEBS J. 2022. PMID: 34145975 Free PMC article. Review.
-
JCPyV VP1 Mutations in Progressive MultifocalLeukoencephalopathy: Altering Tropismor Mediating Immune Evasion?Viruses. 2020 Oct 12;12(10):1156. doi: 10.3390/v12101156. Viruses. 2020. PMID: 33053912 Free PMC article. Review.
-
T cell deficiency precipitates antibody evasion and emergence of neurovirulent polyomavirus.Elife. 2022 Nov 7;11:e83030. doi: 10.7554/eLife.83030. Elife. 2022. PMID: 36341713 Free PMC article.
-
CD8 T Cells and STAT1 Signaling Are Essential Codeterminants in Protection from Polyomavirus Encephalopathy.J Virol. 2020 Mar 31;94(8):e02038-19. doi: 10.1128/JVI.02038-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31996425 Free PMC article.
-
Progressive multifocal leukoencephalopathy-associated mutations in the JC polyomavirus capsid disrupt lactoseries tetrasaccharide c binding.mBio. 2013 Jun 11;4(3):e00247-13. doi: 10.1128/mBio.00247-13. mBio. 2013. PMID: 23760462 Free PMC article.
Cited by
-
Polyomavirus Wakes Up and Chooses Neurovirulence.Viruses. 2023 Oct 18;15(10):2112. doi: 10.3390/v15102112. Viruses. 2023. PMID: 37896889 Free PMC article. Review.
-
Understanding polyomavirus CNS disease - a perspective from mouse models.FEBS J. 2022 Oct;289(19):5744-5761. doi: 10.1111/febs.16083. Epub 2021 Jul 2. FEBS J. 2022. PMID: 34145975 Free PMC article. Review.
-
The CXCR6-CXCL16 axis mediates T cell control of polyomavirus infection in the kidney.PLoS Pathog. 2025 Mar 5;21(3):e1012969. doi: 10.1371/journal.ppat.1012969. eCollection 2025 Mar. PLoS Pathog. 2025. PMID: 40043065 Free PMC article.
-
JCPyV VP1 Mutations in Progressive MultifocalLeukoencephalopathy: Altering Tropismor Mediating Immune Evasion?Viruses. 2020 Oct 12;12(10):1156. doi: 10.3390/v12101156. Viruses. 2020. PMID: 33053912 Free PMC article. Review.
-
Brincidofovir inhibits polyomavirus infection in vivo.mBio. 2024 Aug 14;15(8):e0104924. doi: 10.1128/mbio.01049-24. Epub 2024 Jul 2. mBio. 2024. PMID: 38953354 Free PMC article.
References
-
- Abrishami V, Ilca SL, Gomez-Blanco J, Rissanen I, de la Rosa-Trevín JM, Reddy VS, Carazo JM, Huiskonen JT. Localized reconstruction in Scipion expedites the analysis of symmetry mismatches in cryo-EM data. Progress in Biophysics and Molecular Biology. 2020;S0079-6107:30039-0. doi: 10.1016/j.pbiomolbio.2020.05.00430039-0. - DOI - PubMed
-
- Agnihotri SP, Wuthrich C, Dang X, Nauen D, Karimi R, Viscidi R, Bord E, Batson S, Troncoso J, Koralnik IJ. A fatal case of JC virus meningitis presenting with hydrocephalus in a human immunodeficiency virus-seronegative patient. Annals of Neurology. 2014;76:140–147. doi: 10.1002/ana.24192. - DOI - PMC - PubMed
-
- Asarnow D, Palovcak E, Cheng Y. asarnow/pyem: UCSF pyem. 0.5Zenodo. 2019 doi: 10.5281/zenodo.3576630. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

