Epigenetic silencing of clustered tRNA genes in Arabidopsis
- PMID: 32941623
- PMCID: PMC7544208
- DOI: 10.1093/nar/gkaa766
Epigenetic silencing of clustered tRNA genes in Arabidopsis
Abstract
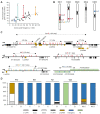
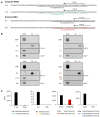
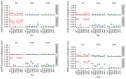
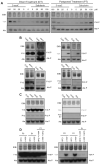
Beyond their key role in translation, cytosolic transfer RNAs (tRNAs) are involved in a wide range of other biological processes. Nuclear tRNA genes (tDNAs) are transcribed by the RNA polymerase III (RNAP III) and cis-elements, trans-factors as well as genomic features are known to influence their expression. In Arabidopsis, besides a predominant population of dispersed tDNAs spread along the 5 chromosomes, some clustered tDNAs have been identified. Here, we demonstrate that these tDNA clusters are transcriptionally silent and that pathways involved in the maintenance of DNA methylation play a predominant role in their repression. Moreover, we show that clustered tDNAs exhibit repressive chromatin features whilst their dispersed counterparts contain permissive euchromatic marks. This work demonstrates that both genomic and epigenomic contexts are key players in the regulation of tDNAs transcription. The conservation of most of these regulatory processes suggests that this pioneering work in Arabidopsis can provide new insights into the regulation of RNA Pol III transcription in other organisms, including vertebrates.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
A unique nucleosome arrangement, maintained actively by chromatin remodelers facilitates transcription of yeast tRNA genes.BMC Genomics. 2013 Jun 17;14:402. doi: 10.1186/1471-2164-14-402. BMC Genomics. 2013. PMID: 23767421 Free PMC article.
-
Transcription of cloned tRNA genes and the nuclear partitioning of a tRNA precursor.Cell. 1979 Dec;18(4):1165-72. doi: 10.1016/0092-8674(79)90229-0. Cell. 1979. PMID: 391407
-
TFIIIC-based chromatin insulators through eukaryotic evolution.Gene. 2022 Aug 15;835:146533. doi: 10.1016/j.gene.2022.146533. Epub 2022 May 24. Gene. 2022. PMID: 35623477
-
Regulation of tRNA gene transcription by the chromatin structure and nucleosome dynamics.Biochim Biophys Acta Gene Regul Mech. 2018 Apr;1861(4):295-309. doi: 10.1016/j.bbagrm.2017.11.008. Epub 2017 Dec 5. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29313808 Review.
-
Extra-transcriptional functions of RNA Polymerase III complexes: TFIIIC as a potential global chromatin bookmark.Gene. 2012 Feb 10;493(2):169-75. doi: 10.1016/j.gene.2011.09.018. Epub 2011 Oct 1. Gene. 2012. PMID: 21986035 Review.
Cited by
-
Genome-wide detection of cytosine methylations in plant from Nanopore data using deep learning.Nat Commun. 2021 Oct 13;12(1):5976. doi: 10.1038/s41467-021-26278-9. Nat Commun. 2021. PMID: 34645826 Free PMC article.
-
tRNA gene content, structure, and organization in the flowering plant lineage.Front Plant Sci. 2024 Dec 23;15:1486612. doi: 10.3389/fpls.2024.1486612. eCollection 2024. Front Plant Sci. 2024. PMID: 39764226 Free PMC article.
-
Efficient endogenous protein labelling in Dictyostelium using CRISPR/Cas9 knock-in and split fluorescent proteins.PLoS One. 2025 Jun 20;20(6):e0326577. doi: 10.1371/journal.pone.0326577. eCollection 2025. PLoS One. 2025. PMID: 40540504 Free PMC article.
-
Epigenetic regulation of functional candidate genes for milk production traits in dairy sheep subjected to protein restriction in the prepubertal stage.BMC Genomics. 2023 Sep 1;24(1):511. doi: 10.1186/s12864-023-09611-y. BMC Genomics. 2023. PMID: 37658326 Free PMC article.
-
Intragenic tRNA-promoted R-loops orchestrate transcription interference for plant oxidative stress responses.Plant Cell. 2021 Nov 4;33(11):3574-3591. doi: 10.1093/plcell/koab220. Plant Cell. 2021. PMID: 34463741 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

