DGINN, an automated and highly-flexible pipeline for the detection of genetic innovations on protein-coding genes
- PMID: 32941639
- PMCID: PMC7544217
- DOI: 10.1093/nar/gkaa680
DGINN, an automated and highly-flexible pipeline for the detection of genetic innovations on protein-coding genes
Abstract
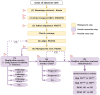
Adaptive evolution has shaped major biological processes. Finding the protein-coding genes and the sites that have been subjected to adaptation during evolutionary time is a major endeavor. However, very few methods fully automate the identification of positively selected genes, and widespread sources of genetic innovations such as gene duplication and recombination are absent from most pipelines. Here, we developed DGINN, a highly-flexible and public pipeline to Detect Genetic INNovations and adaptive evolution in protein-coding genes. DGINN automates, from a gene's sequence, all steps of the evolutionary analyses necessary to detect the aforementioned innovations, including the search for homologs in databases, assignation of orthology groups, identification of duplication and recombination events, as well as detection of positive selection using five methods to increase precision and ranking of genes when a large panel is analyzed. DGINN was validated on nineteen genes with previously-characterized evolutionary histories in primates, including some engaged in host-pathogen arms-races. Our results confirm and also expand results from the literature, including novel findings on the Guanylate-binding protein family, GBPs. This establishes DGINN as an efficient tool to automatically detect genetic innovations and adaptive evolution in diverse datasets, from the user's gene of interest to a large gene list in any species range.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Host-Virus Arms Races Drive Elevated Adaptive Evolution in Viral Receptors.J Virol. 2020 Jul 30;94(16):e00684-20. doi: 10.1128/JVI.00684-20. Print 2020 Jul 30. J Virol. 2020. PMID: 32493827 Free PMC article.
-
Distinct evolutionary trajectories of SARS-CoV-2-interacting proteins in bats and primates identify important host determinants of COVID-19.Proc Natl Acad Sci U S A. 2022 Aug 30;119(35):e2206610119. doi: 10.1073/pnas.2206610119. Epub 2022 Aug 10. Proc Natl Acad Sci U S A. 2022. PMID: 35947637 Free PMC article.
-
Diverse selective regimes shape genetic diversity at ADAR genes and at their coding targets.RNA Biol. 2015;12(2):149-61. doi: 10.1080/15476286.2015.1017215. RNA Biol. 2015. PMID: 25826567 Free PMC article.
-
Patterns of evolution of host proteins involved in retroviral pathogenesis.Retrovirology. 2006 Feb 7;3:11. doi: 10.1186/1742-4690-3-11. Retrovirology. 2006. PMID: 16460575 Free PMC article. Review.
-
Nonadaptive processes in primate and human evolution.Am J Phys Anthropol. 2010;143 Suppl 51:13-45. doi: 10.1002/ajpa.21439. Am J Phys Anthropol. 2010. PMID: 21086525 Review.
Cited by
-
AOC: Analysis of Orthologous Collections - an application for the characterization of natural selection in protein-coding sequences.ArXiv [Preprint]. 2024 Jun 13:arXiv:2406.09522v1. ArXiv. 2024. PMID: 38947939 Free PMC article. Preprint.
-
Understanding the evolution of immune genes in jawed vertebrates.J Evol Biol. 2023 Jun;36(6):847-873. doi: 10.1111/jeb.14181. Epub 2023 May 31. J Evol Biol. 2023. PMID: 37255207 Free PMC article. Review.
-
Evolutionary immunology to explore original antiviral strategies.Philos Trans R Soc Lond B Biol Sci. 2024 May 6;379(1901):20230068. doi: 10.1098/rstb.2023.0068. Epub 2024 Mar 18. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 38497262 Free PMC article. Review.
-
Rapid Evolution of HERC6 and Duplication of a Chimeric HERC5/6 Gene in Rodents and Bats Suggest an Overlooked Role of HERCs in Mammalian Immunity.Front Immunol. 2020 Dec 18;11:605270. doi: 10.3389/fimmu.2020.605270. eCollection 2020. Front Immunol. 2020. PMID: 33391270 Free PMC article.
-
FREEDA: an automated computational pipeline guides experimental testing of protein innovation by detecting positive selection.bioRxiv [Preprint]. 2023 Feb 28:2023.02.27.530329. doi: 10.1101/2023.02.27.530329. bioRxiv. 2023. Update in: J Cell Biol. 2023 Sep 4;222(9):e202212084. doi: 10.1083/jcb.202212084. PMID: 36909479 Free PMC article. Updated. Preprint.
References
-
- Daugherty M.D., Malik H.S.. Rules of engagement: molecular insights from host-virus arms races. Annu. Rev. Genet. 2012; 46:677–700. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

