Human B Cell Clonal Expansion and Convergent Antibody Responses to SARS-CoV-2
- PMID: 32941787
- PMCID: PMC7470783
- DOI: 10.1016/j.chom.2020.09.002
Human B Cell Clonal Expansion and Convergent Antibody Responses to SARS-CoV-2
Abstract
B cells are critical for the production of antibodies and protective immunity to viruses. Here we show that patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) who develop coronavirus disease 2019 (COVID-19) display early recruitment of B cells expressing a limited subset of IGHV genes, progressing to a highly polyclonal response of B cells with broader IGHV gene usage and extensive class switching to IgG and IgA subclasses with limited somatic hypermutation in the initial weeks of infection. We identify convergence of antibody sequences across SARS-CoV-2-infected patients, highlighting stereotyped naive responses to this virus. Notably, sequence-based detection in COVID-19 patients of convergent B cell clonotypes previously reported in SARS-CoV infection predicts the presence of SARS-CoV/SARS-CoV-2 cross-reactive antibody titers specific for the receptor-binding domain. These findings offer molecular insights into shared features of human B cell responses to SARS-CoV-2 and SARS-CoV.
Keywords: B cells; COVID-19; SARS-CoV-2; antibodies; antibody repertoire; clonal expansion; convergent antibody response; immunogenetics; immunology; primary infection.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests A.T.S. is a scientific founder of Immunai and receives research funding from Arsenal Biosciences not related to this study. M.H. is an employee of ATUM. S.D.B. has consulted for Regeneron, Sanofi, and Novartis on topics unrelated to this study. S.D.B., K.R., P.S.K., and A.E.P. have filed provisional patent applications related to serological tests for SARS-CoV-2 antibodies. K.C.N. reports grants from National Institute of Allergy and Infectious Diseases (NIAID), Food Allergy Research & Education (FARE), and End Allergies Together (EAT); National Heart, Lung, and Blood Institute (NHLBI), and National Institute of Environmental Health Sciences (NIEHS), and is the director of FARE and World Allergy Organization (WAO) Center of Excellence at Stanford; advisor at Cour Pharma; co-founder of Before Brands, Alladapt, Latitude, and IgGenix; National Scientific Committee member at Immune Tolerance Network (ITN) and National Institutes of Health (NIH); a recipient of a Research Sponsorship from Nestle; Consultant and Advisory Board Member at Before Brands, Alladapt, Iggenix, NHLBI, and Probio; Data and Safety Monitoring Board member at NHLBI; and has US patents for basophil testing, multifood immunotherapy and prevention, monoclonal antibodies from plasmablasts, and devices for diagnostics. The remaining authors declare that they have no competing interests.
Figures
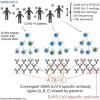


Update of
-
Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2.bioRxiv [Preprint]. 2020 Jul 9:2020.07.08.194456. doi: 10.1101/2020.07.08.194456. bioRxiv. 2020. Update in: Cell Host Microbe. 2020 Oct 7;28(4):516-525.e5. doi: 10.1016/j.chom.2020.09.002. PMID: 32676593 Free PMC article. Updated. Preprint.
-
B cell clonal expansion and convergent antibody responses to SARS-CoV-2.Res Sq [Preprint]. 2020 May 6:rs.3.rs-27220. doi: 10.21203/rs.3.rs-27220/v1. Res Sq. 2020. Update in: Cell Host Microbe. 2020 Oct 7;28(4):516-525.e5. doi: 10.1016/j.chom.2020.09.002. PMID: 32702737 Free PMC article. Updated. Preprint.
Comment in
-
Mining the Antibody Repertoire for Solutions to SARS-CoV-2.Cell Host Microbe. 2020 Oct 7;28(4):499-501. doi: 10.1016/j.chom.2020.09.010. Cell Host Microbe. 2020. PMID: 33031765 Free PMC article.
References
-
- Coughlin M., Lou G., Martinez O., Masterman S.K., Olsen O.A., Moksa A.A., Farzan M., Babcook J.S., Prabhakar B.S. Generation and characterization of human monoclonal neutralizing antibodies with distinct binding and sequence features against SARS coronavirus using XenoMouse. Virology. 2007;361:93–102. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- RM1 HG007735/HG/NHGRI NIH HHS/United States
- T32 AI007502/AI/NIAID NIH HHS/United States
- U54 CA260517/CA/NCI NIH HHS/United States
- K23 HL125663/HL/NHLBI NIH HHS/United States
- DP1 DA046089/DA/NIDA NIH HHS/United States
- K08 CA230188/CA/NCI NIH HHS/United States
- R01 AI127877/AI/NIAID NIH HHS/United States
- U19 AI111825/AI/NIAID NIH HHS/United States
- R01 AI139119/AI/NIAID NIH HHS/United States
- R01 AI130398/AI/NIAID NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- R01 AI125567/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

