Removing reference bias and improving indel calling in ancient DNA data analysis by mapping to a sequence variation graph
- PMID: 32943086
- PMCID: PMC7499850
- DOI: 10.1186/s13059-020-02160-7
Removing reference bias and improving indel calling in ancient DNA data analysis by mapping to a sequence variation graph
Abstract
Background: During the last decade, the analysis of ancient DNA (aDNA) sequence has become a powerful tool for the study of past human populations. However, the degraded nature of aDNA means that aDNA molecules are short and frequently mutated by post-mortem chemical modifications. These features decrease read mapping accuracy and increase reference bias, in which reads containing non-reference alleles are less likely to be mapped than those containing reference alleles. Alternative approaches have been developed to replace the linear reference with a variation graph which includes known alternative variants at each genetic locus. Here, we evaluate the use of variation graph software vg to avoid reference bias for aDNA and compare with existing methods.
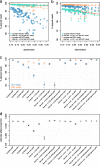
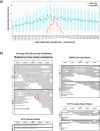
Results: We use vg to align simulated and real aDNA samples to a variation graph containing 1000 Genome Project variants and compare with the same data aligned with bwa to the human linear reference genome. Using vg leads to a balanced allelic representation at polymorphic sites, effectively removing reference bias, and more sensitive variant detection in comparison with bwa, especially for insertions and deletions (indels). Alternative approaches that use relaxed bwa parameter settings or filter bwa alignments can also reduce bias but can have lower sensitivity than vg, particularly for indels.
Conclusions: Our findings demonstrate that aligning aDNA sequences to variation graphs effectively mitigates the impact of reference bias when analyzing aDNA, while retaining mapping sensitivity and allowing detection of variation, in particular indel variation, that was previously missed.
Keywords: Ancient DNA; Reference bias; Sequence alignment; Variation graph.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Brunson K, Reich D. The promise of paleogenomics beyond our own species. Trends Genet. 2019. 10.1016/j.tig.2019.02.006. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

