RNA-dependent RNA polymerase: Structure, mechanism, and drug discovery for COVID-19
- PMID: 32943188
- PMCID: PMC7473028
- DOI: 10.1016/j.bbrc.2020.08.116
RNA-dependent RNA polymerase: Structure, mechanism, and drug discovery for COVID-19
Abstract
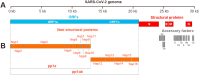
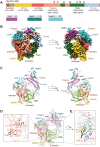
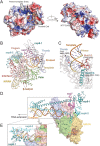
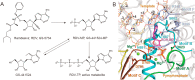
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has rapidly become a global pandemic. Although great efforts have been made to develop effective therapeutic interventions, only the nucleotide analog remdesivir was approved for emergency use against COVID-19. Remdesivir targets the RNA-dependent RNA polymerase (RdRp), an essential enzyme for viral RNA replication and a promising drug target for COVID-19. Recently, several structures of RdRp in complex with substrate RNA and remdesivir were reported, providing insights into the mechanisms of RNA recognition by RdRp. These structures also reveal the mechanism of RdRp inhibition by nucleotide inhibitors and offer a molecular template for the development of RdRp-targeting drugs. This review discusses the recognition mechanism of RNA and nucleotide inhibitor by RdRp, and its implication in drug discovery.
Keywords: COVID-19; RNA recognition; RNA-dependent RNA polymerase; Remdesivir; SARS-CoV-2; Structure.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
-
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265. doi: 10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
-
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., Li W.J., Jiang B.G., Wei W., Yuan T.T., Zheng K., Cui X.M., Li J., Pei G.Q., Qiang X., Cheung W.Y., Li L.F., Sun F.F., Qin S., Huang J.C., Leung G.M., Holmes E.C., Hu Y.L., Guan Y., Cao W.C. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

