Deep amplicon sequencing for culture-free prediction of susceptibility or resistance to 13 anti-tuberculous drugs
- PMID: 32943401
- PMCID: PMC8174722
- DOI: 10.1183/13993003.02338-2020
Deep amplicon sequencing for culture-free prediction of susceptibility or resistance to 13 anti-tuberculous drugs
Erratum in
-
"Deep amplicon sequencing for culture-free prediction of susceptibility or resistance to 13 anti-tuberculous drugs." A. Jouet, C. Gaudin, N. Badalato, et al. Eur Respir J 2021; 57: 2002338.Eur Respir J. 2022 Nov 10;60(5):2052338. doi: 10.1183/13993003.52338-2020. Print 2022 Nov. Eur Respir J. 2022. PMID: 36356977 Free PMC article. No abstract available.
Abstract
Conventional molecular tests for detecting Mycobacterium tuberculosis complex (MTBC) drug resistance on clinical samples cover a limited set of mutations. Whole-genome sequencing (WGS) typically requires culture.Here, we evaluated the Deeplex Myc-TB targeted deep-sequencing assay for prediction of resistance to 13 anti-tuberculous drugs/drug classes, directly applicable on sputum.With MTBC DNA tests, the limit of detection was 100-1000 genome copies for fixed resistance mutations. Deeplex Myc-TB captured in silico 97.1-99.3% of resistance phenotypes correctly predicted by WGS from 3651 MTBC genomes. On 429 isolates, the assay predicted 92.2% of 2369 first- and second-line phenotypes, with a sensitivity of 95.3% and a specificity of 97.4%. 56 out of 69 (81.2%) residual discrepancies with phenotypic results involved pyrazinamide, ethambutol and ethionamide, and low-level rifampicin or isoniazid resistance mutations, all notoriously prone to phenotypic testing variability. Only two out of 91 (2.2%) resistance phenotypes undetected by Deeplex Myc-TB had known resistance-associated mutations by WGS analysis outside Deeplex Myc-TB targets. Phenotype predictions from Deeplex Myc-TB analysis directly on 109 sputa from a Djibouti survey matched those of MTBSeq/PhyResSE/Mykrobe, fed with WGS data from subsequent cultures, with a sensitivity of 93.5/98.5/93.1% and a specificity of 98.5/97.2/95.3%, respectively. Most residual discordances involved gene deletions/indels and 3-12% heteroresistant calls undetected by WGS analysis or natural pyrazinamide resistance of globally rare "Mycobacterium canettii" strains then unreported by Deeplex Myc-TB. On 1494 arduous sputa from a Democratic Republic of the Congo survey, 14 902 out of 19 422 (76.7%) possible susceptible or resistance phenotypes could be predicted culture-free.Deeplex Myc-TB may enable fast, tailored tuberculosis treatment.
Copyright ©ERS 2021.
Conflict of interest statement
Conflict of interest: A. Jouet is an employee of GenoScreen. Conflict of interest: C. Gaudin is an employee of GenoScreen. Conflict of interest: N. Badalato is an employee of GenoScreen. Conflict of interest: C. Allix-Béguec is an employee of GenoScreen. Conflict of interest: S. Duthoy is an employee of GenoScreen. Conflict of interest: A. Ferré is an employee of GenoScreen. Conflict of interest: M. Diels has nothing to disclose. Conflict of interest: Y. Laurent is an employee of GenoScreen. Conflict of interest: S. Contreras is an employee of GenoScreen. Conflict of interest: S. Feuerriegel has nothing to disclose. Conflict of interest: S. Niemann reports grants from the German Center for Infection Research, Excellenz Cluster Precision Medicine in Chronic Inflammation EXC 2167, Leibniz Science Campus Evolutionary Medicine of the LUNG (EvoLUNG) and ECDC public tender: OJ/2017/OCS/7766 “Pilot study on the use of Whole Genome Sequencing for molecular typing and characterisation of M. tuberculosis in the EU/EEA”, during the conduct of the study. Conflict of interest: E. André has nothing to disclose. Conflict of interest: M.K. Kaswa has nothing to disclose. Conflict of interest: E. Tagliani has nothing to disclose. Conflict of interest: A. Cabibbe has nothing to disclose. Conflict of interest: V. Mathys has nothing to disclose. Conflict of interest: D. Cirillo has nothing to disclose. Conflict of interest: B.C. de Jong has nothing to disclose. Conflict of interest: L. Rigouts has nothing to disclose. Conflict of interest: P. Supply reports personal fees for consultancy from GenoScreen, grants from the European Union PathoNGen-Trace project (FP7-278864), during the conduct of the study.
Figures
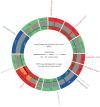
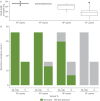
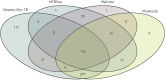
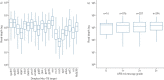
Comment in
-
Targeted next-generation sequencing: a Swiss army knife for mycobacterial diagnostics?Eur Respir J. 2021 Mar 18;57(3):2004077. doi: 10.1183/13993003.04077-2020. Print 2021 Mar. Eur Respir J. 2021. PMID: 33737379 Free PMC article. No abstract available.
References
-
- World Health Organization . Global Tuberculosis Report. Geneva, WHO, 2019.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
