Species-specific pace of development is associated with differences in protein stability
- PMID: 32943498
- PMCID: PMC7116327
- DOI: 10.1126/science.aba7667
Species-specific pace of development is associated with differences in protein stability
Abstract
Although many molecular mechanisms controlling developmental processes are evolutionarily conserved, the speed at which the embryo develops can vary substantially between species. For example, the same genetic program, comprising sequential changes in transcriptional states, governs the differentiation of motor neurons in mouse and human, but the tempo at which it operates differs between species. Using in vitro directed differentiation of embryonic stem cells to motor neurons, we show that the program runs more than twice as fast in mouse as in human. This is not due to differences in signaling, nor the genomic sequence of genes or their regulatory elements. Instead, there is an approximately two-fold increase in protein stability and cell cycle duration in human cells compared with mouse cells. This can account for the slower pace of human development and suggests that differences in protein turnover play a role in interspecies differences in developmental tempo.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures

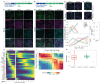

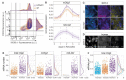
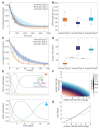

Comment in
-
Tempus fugit: How time flies during development.Science. 2020 Sep 18;369(6510):1431-1432. doi: 10.1126/science.abe0953. Science. 2020. PMID: 32943512 No abstract available.
References
-
- van den Ameele J, Tiberi L, Vanderhaeghen P, Espuny-Camacho I. Thinking out of the dish: what to learn about cortical development using pluripotent stem cells. Trends in Neurosciences. 2014;37:334–342. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

