Single-cell RNA-seq identifies unique transcriptional landscapes of human nucleus pulposus and annulus fibrosus cells
- PMID: 32943704
- PMCID: PMC7499307
- DOI: 10.1038/s41598-020-72261-7
Single-cell RNA-seq identifies unique transcriptional landscapes of human nucleus pulposus and annulus fibrosus cells
Abstract
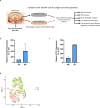
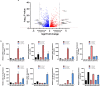
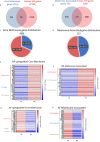
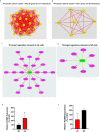
Intervertebral disc (IVD) disease (IDD) is a complex, multifactorial disease. While various aspects of IDD progression have been reported, the underlying molecular pathways and transcriptional networks that govern the maintenance of healthy nucleus pulposus (NP) and annulus fibrosus (AF) have not been fully elucidated. We defined the transcriptome map of healthy human IVD by performing single-cell RNA-sequencing (scRNA-seq) in primary AF and NP cells isolated from non-degenerated lumbar disc. Our systematic and comprehensive analyses revealed distinct genetic architecture of human NP and AF compartments and identified 2,196 differentially expressed genes. Gene enrichment analysis showed that SFRP1, BIRC5, CYTL1, ESM1 and CCNB2 genes were highly expressed in the AF cells; whereas, COL2A1, DSC3, COL9A3, COL11A1, and ANGPTL7 were mostly expressed in the NP cells. Further, functional annotation clustering analysis revealed the enrichment of receptor signaling pathways genes in AF cells, while NP cells showed high expression of genes related to the protein synthesis machinery. Subsequent interaction network analysis revealed a structured network of extracellular matrix genes in NP compartments. Our regulatory network analysis identified FOXM1 and KDM4E as signature transcription factor of AF and NP respectively, which might be involved in the regulation of core genes of AF and NP transcriptome.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Shared and Compartment-Specific Processes in Nucleus Pulposus and Annulus Fibrosus During Intervertebral Disc Degeneration.Adv Sci (Weinh). 2024 May;11(17):e2309032. doi: 10.1002/advs.202309032. Epub 2024 Feb 25. Adv Sci (Weinh). 2024. PMID: 38403470 Free PMC article.
-
Transcriptional profiling of the murine intervertebral disc and age-associated changes in the nucleus pulposus.Connect Tissue Res. 2020 Jan;61(1):63-81. doi: 10.1080/03008207.2019.1665034. Epub 2019 Oct 10. Connect Tissue Res. 2020. PMID: 31597481
-
Single-cell RNA-sequencing atlas of bovine caudal intervertebral discs: Discovery of heterogeneous cell populations with distinct roles in homeostasis.FASEB J. 2021 Nov;35(11):e21919. doi: 10.1096/fj.202101149R. FASEB J. 2021. PMID: 34591994 Free PMC article.
-
The Influence of Axial Compression on the Cellular and Mechanical Function of Spinal Tissues; Emphasis on the Nucleus Pulposus and Annulus Fibrosus: A Review.J Biomech Eng. 2021 May 1;143(5):050802. doi: 10.1115/1.4049749. J Biomech Eng. 2021. PMID: 33454730 Review.
-
Regeneration in Spinal Disease: Therapeutic Role of Hypoxia-Inducible Factor-1 Alpha in Regeneration of Degenerative Intervertebral Disc.Int J Mol Sci. 2021 May 17;22(10):5281. doi: 10.3390/ijms22105281. Int J Mol Sci. 2021. PMID: 34067899 Free PMC article. Review.
Cited by
-
Serglycin secreted by late-stage nucleus pulposus cells is a biomarker of intervertebral disc degeneration.Nat Commun. 2024 Jan 2;15(1):47. doi: 10.1038/s41467-023-44313-9. Nat Commun. 2024. PMID: 38167807 Free PMC article.
-
Revealing the Key MSCs Niches and Pathogenic Genes in Influencing CEP Homeostasis: A Conjoint Analysis of Single-Cell and WGCNA.Front Immunol. 2022 Jun 27;13:933721. doi: 10.3389/fimmu.2022.933721. eCollection 2022. Front Immunol. 2022. PMID: 35833124 Free PMC article.
-
Extracellular matrix gene expression signatures as cell type and cell state identifiers.Matrix Biol Plus. 2021 May 21;10:100069. doi: 10.1016/j.mbplus.2021.100069. eCollection 2021 Jun. Matrix Biol Plus. 2021. PMID: 34195598 Free PMC article.
-
Gene Expression Changes Precede Elevated Mechanical Sensitivity in the Mouse Intervertebral Disc Injury Model.JOR Spine. 2025 Feb 23;8(1):e70049. doi: 10.1002/jsp2.70049. eCollection 2025 Mar. JOR Spine. 2025. PMID: 39989623 Free PMC article.
-
Insights into chondrocyte populations in cartilaginous tissues at the single-cell level.Nat Rev Rheumatol. 2025 Aug;21(8):465-477. doi: 10.1038/s41584-025-01275-0. Epub 2025 Jul 10. Nat Rev Rheumatol. 2025. PMID: 40640375 Review.
References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

