Robust estimation of bacterial cell count from optical density
- PMID: 32943734
- PMCID: PMC7499192
- DOI: 10.1038/s42003-020-01127-5
Robust estimation of bacterial cell count from optical density
Erratum in
-
Author Correction: Robust estimation of bacterial cell count from optical density.Commun Biol. 2020 Oct 27;3(1):640. doi: 10.1038/s42003-020-01371-9. Commun Biol. 2020. PMID: 33110148 Free PMC article.
Abstract
Optical density (OD) is widely used to estimate the density of cells in liquid culture, but cannot be compared between instruments without a standardized calibration protocol and is challenging to relate to actual cell count. We address this with an interlaboratory study comparing three simple, low-cost, and highly accessible OD calibration protocols across 244 laboratories, applied to eight strains of constitutive GFP-expressing E. coli. Based on our results, we recommend calibrating OD to estimated cell count using serial dilution of silica microspheres, which produces highly precise calibration (95.5% of residuals <1.2-fold), is easily assessed for quality control, also assesses instrument effective linear range, and can be combined with fluorescence calibration to obtain units of Molecules of Equivalent Fluorescein (MEFL) per cell, allowing direct comparison and data fusion with flow cytometry measurements: in our study, fluorescence per cell measurements showed only a 1.07-fold mean difference between plate reader and flow cytometry data.
Conflict of interest statement
The authors declare no competing interests.
Figures
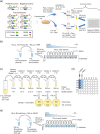
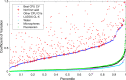



References
-
- DeBlois R, Bean C. Counting and sizing of submicron particles by the resistive pulse technique. Rev. Sci. Instrum. 1970;41:909–916. doi: 10.1063/1.1684724. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

