Competing endogenous RNA network analysis for screening inflammation‑related long non‑coding RNAs for acute ischemic stroke
- PMID: 32945445
- PMCID: PMC7453507
- DOI: 10.3892/mmr.2020.11415
Competing endogenous RNA network analysis for screening inflammation‑related long non‑coding RNAs for acute ischemic stroke
Abstract
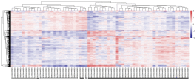
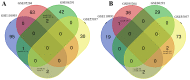
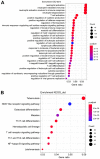
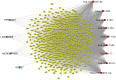
Long non‑coding RNAs (lncRNAs) represent potential biomarkers for the diagnosis and treatment of various diseases; however, the role of circulating acute ischemic stroke (AIS)‑related lncRNAs remains relatively unknown. The present study aimed to screen crucial lncRNAs for AIS based on the competing endogenous RNA (ceRNA) hypothesis. The expression profile datasets for one mRNA, accession no. GSE16561, and four microRNAs (miRNAs), accession nos. GSE95204, GSE86291, GSE55937 and GSE110993, were downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs), lncRNAs (DELs), and miRNAs (DEMs) were identified, and ClusterProfiler was used to interpret the function of the DEGs. Based on the protein‑protein interaction (PPI) network and module analyses, hub DEGs were identified. A ceRNA network was established based on miRNA‑mRNA or miRNA‑lncRNA interaction pairs. In total, 2,041 DEGs and 5 DELs were identified between the AIS and controls samples in GSE16561, and 10 DEMs between at least two of the four miRNA expression profiles. A PPI network was constructed with 1,235 DEGs, among which 20 genes were suggested to be hub genes. The hub genes paxillin (PXN), FYN‑proto‑oncogene, Src family tyrosine kinase (FYN), ras homolog family member A (RHOA), STAT1, and growth factor receptor‑bound protein 2 (GRB2), were amongst the most significantly enriched modules extracted from the PPI network. Functional analysis revealed that these hub genes were associated with inflammation‑related signaling pathways. An AIS‑related ceRNA network was constructed, in which 4 DELs were predicted to function as ceRNAs for 9 DEMs, to regulate the five identified hub genes; that is, minichromosome maintenance complex component 3 associated protein‑antisense RNA 1 (MCM3AP‑AS1)/long intergenic non‑protein coding RNA 1089 (LINC01089)/hsa‑miRNA (miR)‑125a/FYN, inositol‑tetrakisphosphate 1‑kinase‑antisense RNA 1 (ITPK1‑AS1)/hsa‑let‑7i/RHOA/GRB2/STAT1, and human leukocyte antigen complex group 27 (HCG27)/hsa‑-miR‑19a/PXN interaction axes. In conclusion, MCM3AP‑AS1, LINC01089, ITPK1‑AS1, and HCG27 may represent new biomarkers and underlying targets for the treatment of AIS.
Figures


References
-
- Feigin VL, Forouzanfar MH, Krishnamurthi R, Mensah GA, Connor M, Bennett DA, Moran AE, Sacco RL, Anderson L, Truelsen T, et al. Global and regional burden of stroke during 1990–2010: Findings from the Global Burden of disease study 2010. Lancet. 2014;383:3081–255. doi: 10.1016/S0140-6736(13)61953-4. - DOI - PMC - PubMed
-
- Boldsen JK, Engedal TS, Pedraza S, Cho TH, Thomalla G, Nighoghossian N, Baron JC, Fiehler J, Østergaard L, Mouridsen K. Better diffusion segmentation in acute ischemic stroke through automatic tree learning anomaly segmentation. Front Neuroinform. 2018;12:21. doi: 10.3389/fninf.2018.00021. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

