Attenuating the Selection of Vancomycin Resistance Among Enterococci through the Development of Peptide-Based Vancomycin Antagonists
- PMID: 32946213
- PMCID: PMC8006779
- DOI: 10.1021/acsinfecdis.0c00319
Attenuating the Selection of Vancomycin Resistance Among Enterococci through the Development of Peptide-Based Vancomycin Antagonists
Abstract
The emergence and spread of multidrug resistant (MDR) pathogens with acquired resistance to almost all available antimicrobial agents has severely threatened the international healthcare community over the last two decades. The last resort antibiotic vancomycin is critical for treatment of several of these pathogens; howeverc vancomycin resistance is spreading due to the undesired accumulation of IV vancomycin in the colon post-treatment. This accumulation exerts selective pressure upon members of the colonic microflora, including Enterococci, which possess vancomycin resistance genes. To ensure the continual effectiveness of vancomycin in the clinical setting by preventing the spread of antibiotic resistance, it is crucial to develop strategies that reduce selective pressure on the colonic microflora while allowing vancomycin to maintain its desired activity at the site of infection. Herein we report that modification of the native l-Lys-d-Ala-d-Ala vancomycin binding site can be used to produce peptides with the ability to competitively bind vancomycin, reducing its activity against susceptible Enterococci. Moreover, several modifications to the N-termini of the native tripeptide have produced compounds with enhanced vancomycin binding activity, including several analogs that were designed to covalently bind vancomycin, thereby acting as suicide inhibitors. Finally, in a mixed culture of susceptible and resistant bacteria, a single lead compound was found to protect high ratios of susceptible bacteria from vancomycin over the course of a week-long period, preventing the selection for vancomycin-resistant Enterococci. These findings demonstrate the ability of these peptides as potential therapeutic adjuvants for counteracting the undesired accumulation of colonic vancomycin, allowing for protection of the colonic microflora.
Keywords: antibiotic resistance; vancomycin; vancomycin-resistant Enterococcus; viability qPCR.
Figures

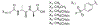
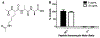











Similar articles
-
Chemical trapping of vancomycin: a potential strategy for preventing selection of vancomycin-resistant Enterococci.Microb Drug Resist. 2012 Apr;18(2):109-15. doi: 10.1089/mdr.2011.0057. Epub 2011 Nov 16. Microb Drug Resist. 2012. PMID: 22088148
-
First Report of the Local Spread of Vancomycin-Resistant Enterococci Ascribed to the Interspecies Transmission of a vanA Gene Cluster-Carrying Linear Plasmid.mSphere. 2020 Apr 8;5(2):e00102-20. doi: 10.1128/mSphere.00102-20. mSphere. 2020. PMID: 32269153 Free PMC article.
-
Poultry as a vector for emerging multidrug resistant Enterococcus spp.: First report of vancomycin (van) and the chloramphenicol-florfenicol (cat-fex-cfr) resistance genes from pigeon and duck faeces.Microb Pathog. 2019 Mar;128:195-205. doi: 10.1016/j.micpath.2019.01.006. Epub 2019 Jan 5. Microb Pathog. 2019. PMID: 30615998
-
Renaissance of vancomycin: approaches for breaking antibiotic resistance in multidrug-resistant bacteria.Can J Microbiol. 2020 Jan;66(1):11-16. doi: 10.1139/cjm-2019-0309. Epub 2019 Sep 23. Can J Microbiol. 2020. PMID: 31545906 Review.
-
Vancomycin-resistant enterococci.Clin Infect Dis. 1998 May;26(5):1196-9. doi: 10.1086/520283. Clin Infect Dis. 1998. PMID: 9597252 Review.
Cited by
-
Polymeric Anti-Antibiotic Microparticles to Prevent Antibiotic Resistance Evolution.Small. 2025 May;21(21):e2407549. doi: 10.1002/smll.202407549. Epub 2025 Jan 19. Small. 2025. PMID: 39828608 Free PMC article.
-
Alternatives to Fight Vancomycin-Resistant Staphylococci and Enterococci.Antibiotics (Basel). 2021 Sep 16;10(9):1116. doi: 10.3390/antibiotics10091116. Antibiotics (Basel). 2021. PMID: 34572698 Free PMC article. Review.
-
The spread of antimicrobial resistance in the aquatic environment from faecal pollution: a scoping review of a multifaceted issue.Environ Monit Assess. 2025 Mar 25;197(4):467. doi: 10.1007/s10661-025-13860-7. Environ Monit Assess. 2025. PMID: 40131552 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

