LSD1 prevents aberrant heterochromatin formation in Neurospora crassa
- PMID: 32946564
- PMCID: PMC7544195
- DOI: 10.1093/nar/gkaa724
LSD1 prevents aberrant heterochromatin formation in Neurospora crassa
Abstract
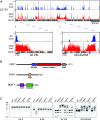
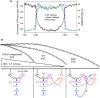
Heterochromatin is a specialized form of chromatin that restricts access to DNA and inhibits genetic processes, including transcription and recombination. In Neurospora crassa, constitutive heterochromatin is characterized by trimethylation of lysine 9 on histone H3, hypoacetylation of histones, and DNA methylation. We explored whether the conserved histone demethylase, lysine-specific demethylase 1 (LSD1), regulates heterochromatin in Neurospora, and if so, how. Though LSD1 is implicated in heterochromatin regulation, its function is inconsistent across different systems; orthologs of LSD1 have been shown to either promote or antagonize heterochromatin expansion by removing H3K4me or H3K9me respectively. We identify three members of the Neurospora LSD complex (LSDC): LSD1, PHF1, and BDP-1. Strains deficient for any of these proteins exhibit variable spreading of heterochromatin and establishment of new heterochromatin domains throughout the genome. Although establishment of H3K9me3 is typically independent of DNA methylation in Neurospora, instances of DNA methylation-dependent H3K9me3 have been found outside regions of canonical heterochromatin. Consistent with this, the hyper-H3K9me3 phenotype of Δlsd1 strains is dependent on the presence of DNA methylation, as well as HCHC-mediated histone deacetylation, suggesting that spreading is dependent on some feedback mechanism. Altogether, our results suggest LSD1 works in opposition to HCHC to maintain proper heterochromatin boundaries.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Induction of H3K9me3 and DNA methylation by tethered heterochromatin factors in Neurospora crassa.Proc Natl Acad Sci U S A. 2017 Nov 7;114(45):E9598-E9607. doi: 10.1073/pnas.1715049114. Epub 2017 Oct 23. Proc Natl Acad Sci U S A. 2017. PMID: 29078403 Free PMC article.
-
DNA methylation and the formation of heterochromatin in Neurospora crassa.Heredity (Edinb). 2010 Jul;105(1):38-44. doi: 10.1038/hdy.2010.44. Epub 2010 Apr 21. Heredity (Edinb). 2010. PMID: 20407471 Review.
-
Histone H3 lysine 4 methyltransferase is required for facultative heterochromatin at specific loci.BMC Genomics. 2019 May 8;20(1):350. doi: 10.1186/s12864-019-5729-7. BMC Genomics. 2019. PMID: 31068130 Free PMC article.
-
Heterochromatin protein 1 forms distinct complexes to direct histone deacetylation and DNA methylation.Nat Struct Mol Biol. 2012 Apr 15;19(5):471-7, S1. doi: 10.1038/nsmb.2274. Nat Struct Mol Biol. 2012. PMID: 22504884 Free PMC article.
-
Caught in conspiracy: cooperation between DNA methylation and histone H3K9 methylation in the establishment and maintenance of heterochromatin.Biochem Cell Biol. 2005 Jun;83(3):385-95. doi: 10.1139/o05-043. Biochem Cell Biol. 2005. PMID: 15959564 Review.
Cited by
-
Regulatory Roles of Histone Modifications in Filamentous Fungal Pathogens.J Fungi (Basel). 2022 May 25;8(6):565. doi: 10.3390/jof8060565. J Fungi (Basel). 2022. PMID: 35736048 Free PMC article. Review.
-
H3K56 deacetylation and H2A.Z deposition are required for aberrant heterochromatin spreading.Nucleic Acids Res. 2022 Apr 22;50(7):3852-3866. doi: 10.1093/nar/gkac196. Nucleic Acids Res. 2022. PMID: 35333354 Free PMC article.
-
Kdm1a safeguards the topological boundaries of PRC2-repressed genes and prevents aging-related euchromatinization in neurons.Nat Commun. 2024 Mar 7;15(1):1781. doi: 10.1038/s41467-024-45773-3. Nat Commun. 2024. PMID: 38453932 Free PMC article.
-
RID is required for both repeat-induced point mutation and nucleation of a novel transitional heterochromatic state for euchromatic repeats.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf263. doi: 10.1093/nar/gkaf263. Nucleic Acids Res. 2025. PMID: 40183634 Free PMC article.
References
-
- Zhang K., Mosch K., Fischle W., Grewal S.I.S.. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 2008; 15:381–388. - PubMed
-
- Coveney J., Woodland H.R.. The DNase I sensitivity of Xenopuslaevis genes transcribed by RNA polymerase III. Nature. 1982; 298:578–580. - PubMed
-
- DeLotto R., Schedl P.. Internal promoter elements of transfer RNA genes are preferentially exposed in chromatin. J. Mol. Biol. 1984; 179:607–628. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

