Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution
- PMID: 32946783
- PMCID: PMC7505495
- DOI: 10.1016/j.cell.2020.08.013
Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution
Abstract
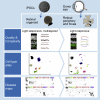
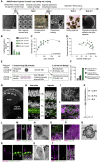
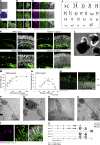
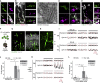
Human organoids recapitulating the cell-type diversity and function of their target organ are valuable for basic and translational research. We developed light-sensitive human retinal organoids with multiple nuclear and synaptic layers and functional synapses. We sequenced the RNA of 285,441 single cells from these organoids at seven developmental time points and from the periphery, fovea, pigment epithelium and choroid of light-responsive adult human retinas, and performed histochemistry. Cell types in organoids matured in vitro to a stable "developed" state at a rate similar to human retina development in vivo. Transcriptomes of organoid cell types converged toward the transcriptomes of adult peripheral retinal cell types. Expression of disease-associated genes was cell-type-specific in adult retina, and cell-type specificity was retained in organoids. We implicate unexpected cell types in diseases such as macular degeneration. This resource identifies cellular targets for studying disease mechanisms in organoids and for targeted repair in human retinas.
Keywords: eye disease; human retina; macular degeneration; organoid; organoid development; retina; retinal organoid; single cell sequencing; synaptic function; transcriptome.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures











References
-
- Aurnhammer C., Haase M., Muether N., Hausl M., Rauschhuber C., Huber I., Nitschko H., Busch U., Sing A., Ehrhardt A., Baiker A. Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum. Gene Ther. Methods. 2012;23:18–28. - PubMed
-
- Baden T., Schubert T., Chang L., Wei T., Zaichuk M., Wissinger B., Euler T. A tale of two retinal domains: near-optimal sampling of achromatic contrasts in natural scenes through asymmetric photoreceptor distribution. Neuron. 2013;80:1206–1217. - PubMed
-
- Balse E., Tessier L.-H., Fuchs C., Forster V., Sahel J.A., Picaud S. Purification of mammalian cone photoreceptors by lectin panning and the enhancement of their survival in glia-conditioned medium. Invest. Ophthalmol. Vis. Sci. 2005;46:367–374. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

