A biaryl sulfonamide derivative as a novel inhibitor of filovirus infection
- PMID: 32946918
- PMCID: PMC11075116
- DOI: 10.1016/j.antiviral.2020.104932
A biaryl sulfonamide derivative as a novel inhibitor of filovirus infection
Abstract
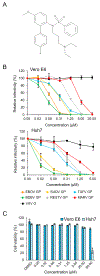
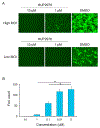
Ebolaviruses and marburgviruses, members of the family Filoviridae, are known to cause fatal diseases often associated with hemorrhagic fever. Recent outbreaks of Ebola virus disease in West African countries and the Democratic Republic of the Congo have made clear the urgent need for the development of therapeutics and vaccines against filoviruses. Using replication-incompetent vesicular stomatitis virus (VSV) pseudotyped with the Ebola virus (EBOV) envelope glycoprotein (GP), we screened a chemical compound library to obtain new drug candidates that inhibit filoviral entry into target cells. We discovered a biaryl sulfonamide derivative that suppressed in vitro infection mediated by GPs derived from all known human-pathogenic filoviruses. To determine the inhibitory mechanism of the compound, we monitored each entry step (attachment, internalization, and membrane fusion) using lipophilic tracer-labeled ebolavirus-like particles and found that the compound efficiently blocked fusion between the viral envelope and the endosomal membrane during cellular entry. However, the compound did not block the interaction of GP with the Niemann-Pick C1 protein, which is believed to be the receptor of filoviruses. Using replication-competent VSVs pseudotyped with EBOV GP, we selected escape mutants and identified two EBOV GP amino acid residues (positions 47 and 66) important for the interaction with this compound. Interestingly, these amino acid residues were located at the base region of the GP trimer, suggesting that the compound might interfere with the GP conformational change required for membrane fusion. These results suggest that this biaryl sulfonamide derivative is a novel fusion inhibitor and a possible drug candidate for the development of a pan-filovirus therapeutic.
Keywords: Compound; Ebolavirus; Entry inhibitor; Glycoprotein; Marburgvirus; Membrane fusion.
Copyright © 2020 Elsevier B.V. All rights reserved.
Figures


Similar articles
-
Identification of filovirus entry inhibitors targeting the endosomal receptor NPC1 binding site.Antiviral Res. 2021 May;189:105059. doi: 10.1016/j.antiviral.2021.105059. Epub 2021 Mar 8. Antiviral Res. 2021. PMID: 33705865 Free PMC article.
-
Inhibition of Ebola and Marburg Virus Entry by G Protein-Coupled Receptor Antagonists.J Virol. 2015 Oct;89(19):9932-8. doi: 10.1128/JVI.01337-15. Epub 2015 Jul 22. J Virol. 2015. PMID: 26202243 Free PMC article.
-
A Diacylglycerol Kinase Inhibitor, R-59-022, Blocks Filovirus Internalization in Host Cells.Viruses. 2019 Mar 1;11(3):206. doi: 10.3390/v11030206. Viruses. 2019. PMID: 30832223 Free PMC article.
-
Current status of small molecule drug development for Ebola virus and other filoviruses.Curr Opin Virol. 2019 Apr;35:42-56. doi: 10.1016/j.coviro.2019.03.001. Epub 2019 Apr 16. Curr Opin Virol. 2019. PMID: 31003196 Free PMC article. Review.
-
Filovirus entry: a novelty in the viral fusion world.Viruses. 2012 Feb;4(2):258-75. doi: 10.3390/v4020258. Epub 2012 Feb 7. Viruses. 2012. PMID: 22470835 Free PMC article. Review.
Cited by
-
Development of an imaging system for visualization of Ebola virus glycoprotein throughout the viral lifecycle.Front Microbiol. 2022 Nov 3;13:1026644. doi: 10.3389/fmicb.2022.1026644. eCollection 2022. Front Microbiol. 2022. PMID: 36406413 Free PMC article.
-
Development of a Novel, Highly Sensitive System for Evaluating Ebola Virus Particle Formation.Viruses. 2025 Jul 19;17(7):1016. doi: 10.3390/v17071016. Viruses. 2025. PMID: 40733632 Free PMC article.
References
-
- Bai CQ, Mu JS, Kargbo D, Song Y. Bin, Niu WK, Nie WM, Kanu A, Liu WW, Wang YP, Dafae F, Yan T, Hu Y, Deng YQ, Lu HJ, Yang F, Zhang XG, Sun Yang, Cao YX, Su HX, Sun Yu, Liu W. Sen, Wang CY, Qian J, Liu L, Wang H, Tong YG, Liu ZY, Chen YS, Wang HQ, Kargbo B, Gao GF, Jiang JF, 2016. Clinical and virological characteristics of ebola virus disease patients treated with favipiravir (T-705) - Sierra Leone, 2014. Clin. Infect. Dis 63, 1288–1294. 10.1093/cid/ciw571. - DOI - PubMed

