Structure, Folding and Stability of Nucleoside Diphosphate Kinases
- PMID: 32947863
- PMCID: PMC7554756
- DOI: 10.3390/ijms21186779
Structure, Folding and Stability of Nucleoside Diphosphate Kinases
Abstract
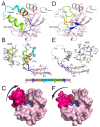
Nucleoside diphosphate kinases (NDPK) are oligomeric proteins involved in the synthesis of nucleoside triphosphates. Their tridimensional structure has been solved by X-ray crystallography and shows that individual subunits present a conserved ferredoxin fold of about 140 residues in prokaryotes, archaea, eukaryotes and viruses. Monomers are functionally independent from each other inside NDPK complexes and the nucleoside kinase catalytic mechanism involves transient phosphorylation of the conserved catalytic histidine. To be active, monomers must assemble into conserved head to tail dimers, which further assemble into hexamers or tetramers. The interfaces between these oligomeric states are very different but, surprisingly, the assembly structure barely affects the catalytic efficiency of the enzyme. While it has been shown that assembly into hexamers induces full formation of the catalytic site and stabilizes the complex, it is unclear why assembly into tetramers is required for function. Several additional activities have been revealed for NDPK, especially in metastasis spreading, cytoskeleton dynamics, DNA binding and membrane remodeling. However, we still lack the high resolution structural data of NDPK in complex with different partners, which is necessary for deciphering the mechanism of these diverse functions. In this review we discuss advances in the structure, folding and stability of NDPKs.
Keywords: histidine kinase; nucleoside diphosphate kinase structure; oligomeric state; protein folding; protein stability; quaternary structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

