In Vitro Characterization of Gut Microbiota-Derived Commensal Strains: Selection of Parabacteroides distasonis Strains Alleviating TNBS-Induced Colitis in Mice
- PMID: 32947881
- PMCID: PMC7565435
- DOI: 10.3390/cells9092104
In Vitro Characterization of Gut Microbiota-Derived Commensal Strains: Selection of Parabacteroides distasonis Strains Alleviating TNBS-Induced Colitis in Mice
Abstract
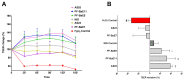
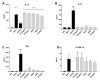
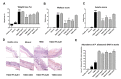
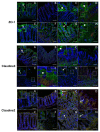
Alterations in the gut microbiota composition and diversity seem to play a role in the development of chronic diseases, including inflammatory bowel disease (IBD), leading to gut barrier disruption and induction of proinflammatory immune responses. This opens the door for the use of novel health-promoting bacteria. We selected five Parabacteroides distasonis strains isolated from human adult and neonates gut microbiota. We evaluated in vitro their immunomodulation capacities and their ability to reinforce the gut barrier and characterized in vivo their protective effects in an acute murine model of colitis. The in vitro beneficial activities were highly strain dependent: two strains exhibited a potent anti-inflammatory potential and restored the gut barrier while a third strain reinstated the epithelial barrier. While their survival to in vitro gastric conditions was variable, the levels of P. distasonis DNA were higher in the stools of bacteria-treated animals. The strains that were positively scored in vitro displayed a strong ability to rescue mice from colitis. We further showed that two strains primed dendritic cells to induce regulatory T lymphocytes from naïve CD4+ T cells. This study provides better insights on the functionality of commensal bacteria and crucial clues to design live biotherapeutics able to target inflammatory chronic diseases such as IBD.
Keywords: IBD; colitis; functional screening; holobiont; immune response; live biotherapeutic products (LBP); microbiota; probiotics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Akkermansia muciniphila and Parabacteroides distasonis synergistically protect from colitis by promoting ILC3 in the gut.mBio. 2024 Apr 10;15(4):e0007824. doi: 10.1128/mbio.00078-24. Epub 2024 Mar 12. mBio. 2024. PMID: 38470269 Free PMC article.
-
New probiotic strains for inflammatory bowel disease management identified by combining in vitro and in vivo approaches.Benef Microbes. 2018 Feb 27;9(2):317-331. doi: 10.3920/BM2017.0097. Benef Microbes. 2018. PMID: 29488412
-
Bacteriocin biosynthesis contributes to the anti-inflammatory capacities of probiotic Lactobacillus plantarum.Benef Microbes. 2018 Feb 27;9(2):333-344. doi: 10.3920/BM2017.0096. Epub 2017 Oct 25. Benef Microbes. 2018. PMID: 29065706
-
Interrelatedness between dysbiosis in the gut microbiota due to immunodeficiency and disease penetrance of colitis.Immunology. 2015 Nov;146(3):359-68. doi: 10.1111/imm.12511. Epub 2015 Sep 7. Immunology. 2015. PMID: 26211540 Free PMC article. Review.
-
Gut bacteria signaling to mitochondria in intestinal inflammation and cancer.Gut Microbes. 2020 May 3;11(3):285-304. doi: 10.1080/19490976.2019.1592421. Epub 2019 Mar 26. Gut Microbes. 2020. PMID: 30913966 Free PMC article. Review.
Cited by
-
Yeast mannoproteins are expected to be a novel potential functional food for attenuation of obesity and modulation of gut microbiota.Front Nutr. 2022 Oct 14;9:1019344. doi: 10.3389/fnut.2022.1019344. eCollection 2022. Front Nutr. 2022. PMID: 36313084 Free PMC article.
-
Akkermansia muciniphila and Parabacteroides distasonis as prognostic markers for relapse in ulcerative colitis patients.Front Cell Infect Microbiol. 2024 Jul 4;14:1367998. doi: 10.3389/fcimb.2024.1367998. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39027140 Free PMC article.
-
The Impact of Escherichia coli Probiotic Strain O83:K24:H31 on the Maturation of Dendritic Cells and Immunoregulatory Functions In Vitro and In Vivo.Cells. 2022 May 12;11(10):1624. doi: 10.3390/cells11101624. Cells. 2022. PMID: 35626660 Free PMC article.
-
Inflammatory Proteins Mediate the Effect of Gut Microbiota on Graves' Ophthalmopathy: A Mendelian Randomization Study.Transl Vis Sci Technol. 2025 Jun 2;14(6):34. doi: 10.1167/tvst.14.6.34. Transl Vis Sci Technol. 2025. PMID: 40576426 Free PMC article.
-
Parabacteroides distasonis Properties Linked to the Selection of New Biotherapeutics.Nutrients. 2022 Oct 7;14(19):4176. doi: 10.3390/nu14194176. Nutrients. 2022. PMID: 36235828 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

