Physiological Approach to the Use of the Natural Compound Quinate in the Control of Sensitive and Resistant Papaver rhoeas
- PMID: 32948013
- PMCID: PMC7569983
- DOI: 10.3390/plants9091215
Physiological Approach to the Use of the Natural Compound Quinate in the Control of Sensitive and Resistant Papaver rhoeas
Abstract
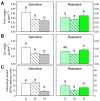
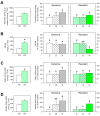
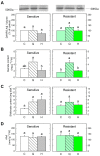
Quinate (1,3,4,5-tetrahydroxycyclohexanecarboxylate) is a compound synthesized in plants through a side-branch of the shikimate biosynthesis pathway, which is accumulated after glyphosate and acetolactate synthase inhibiting herbicides (ALS-inhibitors) and has phytotoxic potential. The objective of this study was to evaluate the phytotoxicity of quinate on several weed species. Among the species evaluated, Cynodon dactylon, Bromus diandrus, Lolium rigidum, Sinapis alba, and Papaver rhoeas, P. rhoeas was the most sensitive, and its growth was controlled with quinate concentrations above 100 mM at the phenological stage of 6-8 true leaves. A physiological study, including the shikimate pathway and the physiological markers of ALS-inhibitors (carbohydrates and amino acids), was performed in the sensitive and resistant plants treated with sulfonylureas or quinate. The typical physiological effects of ALS-inhibitors were detected in the sensitive population (free amino acid and carbohydrate accumulation) and not detected in the resistant population. The mode of action of quinate appeared to be related to general perturbations in their carbon/nitrogen metabolism rather than to specific changes in the shikimate pathway. These results suggest the possibility of using quinate in the weed control management of P. rhoeas.
Keywords: corn poppy; free amino acids; physiological effects; quinate; shikimate pathway; sulfonylureas.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The possible role of quinate in the mode of action of glyphosate and acetolactate synthase inhibitors.Pest Manag Sci. 2010 Mar;66(3):262-9. doi: 10.1002/ps.1868. Pest Manag Sci. 2010. PMID: 19918955
-
The pattern of shikimate pathway and phenylpropanoids after inhibition by glyphosate or quinate feeding in pea roots.Pestic Biochem Physiol. 2017 Sep;141:96-102. doi: 10.1016/j.pestbp.2016.12.005. Epub 2016 Dec 10. Pestic Biochem Physiol. 2017. PMID: 28911748
-
Occurrence, genetic control and evolution of non-target-site based resistance to herbicides inhibiting acetolactate synthase (ALS) in the dicot weed Papaver rhoeas.Plant Sci. 2015 Sep;238:158-69. doi: 10.1016/j.plantsci.2015.06.008. Epub 2015 Jun 12. Plant Sci. 2015. PMID: 26259184
-
Iconic Arable Weeds: The Significance of Corn Poppy (Papaver rhoeas), Cornflower (Centaurea cyanus), and Field Larkspur (Delphinium consolida) in Hungarian Ethnobotanical and Cultural Heritage.Plants (Basel). 2022 Dec 23;12(1):84. doi: 10.3390/plants12010084. Plants (Basel). 2022. PMID: 36616213 Free PMC article. Review.
-
An introduction to ALS-inhibiting herbicides.Toxicol Ind Health. 1999 Jan-Mar;15(1-2):231-9. doi: 10.1191/074823399678846592. Toxicol Ind Health. 1999. PMID: 10188205 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

