Genome-wide identification and expression patterns analysis of the RPD3/HDA1 gene family in cotton
- PMID: 32948145
- PMCID: PMC7501681
- DOI: 10.1186/s12864-020-07069-w
Genome-wide identification and expression patterns analysis of the RPD3/HDA1 gene family in cotton
Abstract
Background: Histone deacetylases (HDACs) catalyze histone deacetylation and suppress gene transcription during various cellular processes. Within the superfamily of HDACs, RPD3/HDA1-type HDACs are the most studied, and it is reported that RPD3 genes play crucial roles in plant growth and physiological processes. However, there is a lack of systematic research on the RPD3/HDA1 gene family in cotton.
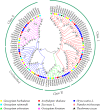
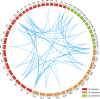
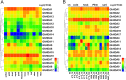
Results: In this study, genome-wide analysis identified 9, 9, 18, and 18 RPD3 genes in Gossypium raimondii, G. arboreum, G. hirsutum, and G. barbadense, respectively. This gene family was divided into 4 subfamilies through phylogenetic analysis. The exon-intron structure and conserved motif analysis revealed high conservation in each branch of the cotton RPD3 genes. Collinearity analysis indicated that segmental duplication was the primary driving force during the expansion of the RPD3 gene family in cotton. There was at least one presumed cis-element related to plant hormones in the promoter regions of all GhRPD3 genes, especially MeJA- and ABA-responsive elements, which have more members than other hormone-relevant elements. The expression patterns showed that most GhRPD3 genes had relatively high expression levels in floral organs and performed higher expression in early-maturity cotton compared with late-maturity cotton during flower bud differentiation. In addition, the expression of GhRPD3 genes could be significantly induced by one or more abiotic stresses as well as exogenous application of MeJA or ABA.
Conclusions: Our findings reveal that GhRPD3 genes may be involved in flower bud differentiation and resistance to abiotic stresses, which provides a basis for further functional verification of GhRPD3 genes in cotton development and a foundation for breeding better early-maturity cotton cultivars in the future.
Keywords: Abiotic stress; Early maturity; Expression patterns; Gossypium; Histone deacetylases.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
Histone Deacetylase (HDAC) Gene Family in Allotetraploid Cotton and Its Diploid Progenitors: In Silico Identification, Molecular Characterization, and Gene Expression Analysis under Multiple Abiotic Stresses, DNA Damage and Phytohormone Treatments.Int J Mol Sci. 2020 Jan 3;21(1):321. doi: 10.3390/ijms21010321. Int J Mol Sci. 2020. PMID: 31947720 Free PMC article.
-
Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses.BMC Genomics. 2018 Aug 9;19(1):599. doi: 10.1186/s12864-018-4985-2. BMC Genomics. 2018. PMID: 30092779 Free PMC article.
-
Genome-wide analyses of member identification, expression pattern, and protein-protein interaction of EPF/EPFL gene family in Gossypium.BMC Plant Biol. 2024 Jun 14;24(1):554. doi: 10.1186/s12870-024-05262-7. BMC Plant Biol. 2024. PMID: 38877405 Free PMC article.
-
Genome-wide expression analysis of phospholipase A1 (PLA1) gene family suggests phospholipase A1-32 gene responding to abiotic stresses in cotton.Int J Biol Macromol. 2021 Dec 1;192:1058-1074. doi: 10.1016/j.ijbiomac.2021.10.038. Epub 2021 Oct 14. Int J Biol Macromol. 2021. PMID: 34656543 Review.
-
Identification of BR biosynthesis genes in cotton reveals that GhCPD-3 restores BR biosynthesis and mediates plant growth and development.Planta. 2021 Sep 17;254(4):75. doi: 10.1007/s00425-021-03727-9. Planta. 2021. PMID: 34533620 Review.
Cited by
-
Genomics, Proteomics, and Metabolomics Approaches to Improve Abiotic Stress Tolerance in Tomato Plant.Int J Mol Sci. 2023 Feb 3;24(3):3025. doi: 10.3390/ijms24033025. Int J Mol Sci. 2023. PMID: 36769343 Free PMC article. Review.
-
Protein S-Acyl Transferase GhPAT27 Was Associated with Verticillium wilt Resistance in Cotton.Plants (Basel). 2022 Oct 18;11(20):2758. doi: 10.3390/plants11202758. Plants (Basel). 2022. PMID: 36297782 Free PMC article.
-
Genome-wide identification and expression profile of YABBY genes in Averrhoa carambola.PeerJ. 2022 Jan 4;9:e12558. doi: 10.7717/peerj.12558. eCollection 2022. PeerJ. 2022. PMID: 35036123 Free PMC article.
-
Genome-Wide Characterization, Evolutionary Analysis of ARF Gene Family, and the Role of SaARF4 in Cd Accumulation of Sedum alfredii Hance.Plants (Basel). 2022 May 9;11(9):1273. doi: 10.3390/plants11091273. Plants (Basel). 2022. PMID: 35567274 Free PMC article.
-
Evolution and Stress Responses of CLO Genes and Potential Function of the GhCLO06 Gene in Salt Resistance of Cotton.Front Plant Sci. 2022 Jan 17;12:801239. doi: 10.3389/fpls.2021.801239. eCollection 2021. Front Plant Sci. 2022. PMID: 35111180 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

