Caffeic acid derivatives (CAFDs) as inhibitors of SARS-CoV-2: CAFDs-based functional foods as a potential alternative approach to combat COVID-19
- PMID: 32948420
- PMCID: PMC7442560
- DOI: 10.1016/j.phymed.2020.153310
Caffeic acid derivatives (CAFDs) as inhibitors of SARS-CoV-2: CAFDs-based functional foods as a potential alternative approach to combat COVID-19
Abstract
Background: SARS-CoV-2, an emerging strain of coronavirus, has affected millions of people from all the continents of world and received worldwide attention. This emerging health crisis calls for the urgent development of specific therapeutics against COVID-19 to potentially reduce the burden of this emerging pandemic.
Purpose: This study aims to evaluate the anti-viral efficacy of natural bioactive entities against COVID-19 via molecular docking and molecular dynamics simulation.
Methods: A library of 27 caffeic-acid derivatives was screened against 5 proteins of SARS-CoV-2 by using Molegro Virtual Docker 7 to obtain the binding energies and interactions between compounds and SARS-CoV-2 proteins. ADME properties and toxicity profiles were investigated via www.swissadme.ch web tools and Toxtree respectively. Molecular dynamics simulation was performed to determine the stability of the lead-protein interactions.
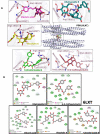
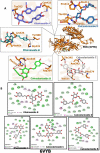
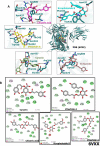
Results: Our obtained results has uncovered khainaoside C, 6-O-Caffeoylarbutin, khainaoside B, khainaoside C and vitexfolin A as potent modulators of COVID-19 possessing more binding energies than nelfinavir against COVID-19 Mpro, Nsp15, SARS-CoV-2 spike S2 subunit, spike open state and closed state structure respectively. While Calceolarioside B was identified as pan inhibitor, showing strong molecular interactions with all proteins except SARS-CoV-2 spike glycoprotein closed state. The results are supported by 20 ns molecular dynamics simulations of the best complexes.
Conclusion: This study will hopefully pave a way for development of phytonutrients-based antiviral therapeutic for treatment or prevention of COVID-19 and further studies are recommended to evaluate the antiviral effects of these phytochemicals against SARS-CoV-2 in in vitro and in vivo models.
Keywords: Bioactive phytochemicals; COVID-19; Caffeic acid derivative; SARS-CoV-2.
Copyright © 2020 Elsevier GmbH. All rights reserved.
Figures







Similar articles
-
Structure-based drug repurposing for targeting Nsp9 replicase and spike proteins of severe acute respiratory syndrome coronavirus 2.J Biomol Struct Dyn. 2022 Jan;40(1):249-262. doi: 10.1080/07391102.2020.1811773. Epub 2020 Aug 24. J Biomol Struct Dyn. 2022. PMID: 32838660 Free PMC article.
-
Phytoconstituents from Moringa oleifera fruits target ACE2 and open spike glycoprotein to combat SARS-CoV-2: An integrative phytochemical and computational approach.J Food Biochem. 2022 May;46(5):e14062. doi: 10.1111/jfbc.14062. Epub 2022 Jan 19. J Food Biochem. 2022. PMID: 35043973
-
Lead Finding from Selected Flavonoids with Antiviral (SARS-CoV-2) Potentials Against COVID-19: An In-silico Evaluation.Comb Chem High Throughput Screen. 2021;24(6):879-890. doi: 10.2174/1386207323999200818162706. Comb Chem High Throughput Screen. 2021. PMID: 32819226
-
Molecular scaffolds from mother nature as possible lead compounds in drug design and discovery against coronaviruses: A landscape analysis of published literature and molecular docking studies.Microb Pathog. 2021 Aug;157:104933. doi: 10.1016/j.micpath.2021.104933. Epub 2021 May 11. Microb Pathog. 2021. PMID: 33984466 Free PMC article. Review.
-
Plant lectins as prospective antiviral biomolecules in the search for COVID-19 eradication strategies.Biomed Pharmacother. 2022 Feb;146:112507. doi: 10.1016/j.biopha.2021.112507. Epub 2021 Dec 7. Biomed Pharmacother. 2022. PMID: 34891122 Free PMC article. Review.
Cited by
-
Promising natural products against SARS-CoV-2: Structure, function, and clinical trials.Phytother Res. 2022 Oct;36(10):3833-3858. doi: 10.1002/ptr.7580. Epub 2022 Aug 5. Phytother Res. 2022. PMID: 35932157 Free PMC article. Review.
-
Efficacy and safety of add-on Viola odorata L. in the treatment of COVID-19: A randomized double-blind controlled trial.J Ethnopharmacol. 2023 Mar 25;304:116058. doi: 10.1016/j.jep.2022.116058. Epub 2022 Dec 17. J Ethnopharmacol. 2023. PMID: 36535329 Free PMC article. Clinical Trial.
-
In Silico Screening of Bioactive Compounds of Representative Seaweeds to Inhibit SARS-CoV-2 ACE2-Bound Omicron B.1.1.529 Spike Protein Trimer.Mar Drugs. 2022 Feb 17;20(2):148. doi: 10.3390/md20020148. Mar Drugs. 2022. PMID: 35200677 Free PMC article.
-
Globularia alypum L. and Related Species: LC-MS Profiles and Antidiabetic, Antioxidant, Anti-Inflammatory, Antibacterial and Anticancer Potential.Pharmaceuticals (Basel). 2022 Apr 21;15(5):506. doi: 10.3390/ph15050506. Pharmaceuticals (Basel). 2022. PMID: 35631332 Free PMC article.
-
Inhibitory Effects and Surface Plasmon Resonance-Based Binding Affinities of Dietary Hydrolyzable Tannins and Their Gut Microbial Metabolites on SARS-CoV-2 Main Protease.J Agric Food Chem. 2021 Oct 20;69(41):12197-12208. doi: 10.1021/acs.jafc.1c03521. Epub 2021 Sep 29. J Agric Food Chem. 2021. PMID: 34586788 Free PMC article.
References
-
- Abu-Reidah I.M., Contreras M.M., Arraez-Roman D., Segura-Carretero A., Fernandez-Gutierrez A. Reversed-phase ultra-high-performance liquid chromatography coupled to electrospray ionization-quadrupole-time-of-flight mass spectrometry as a powerful tool for metabolic profiling of vegetables: Lactuca sativa as an example of its application. J. Chromatogr. A. 2013;1313:212–227. - PubMed
-
- Amoretti M., Amsler C., Bonomi G., Bouchta A., Bowe P., Carraro C., Cesar C.L., Charlton M., Collier M.J., Doser M., Filippini V., Fine K.S., Fontana A., Fujiwara M.C., Funakoshi R., Genova P., Hangst J.S., Hayano R.S., Holzscheiter M.H., Jorgensen L.V., Lagomarsino V., Landua R., Lindelof D., Lodi Rizzini E., Macri M., Madsen N., Manuzio G., Marchesotti M., Montagna P., Pruys H., Regenfus C., Riedler P., Rochet J., Rotondi A., Rouleau G., Testera G., Variola A., Watson T.L., van der Werf D.P., Collaboration A. Production and detection of cold antihydrogen atoms. Nature. 2002;419:456–459. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

