Competence-Stimulating-Peptide-Dependent Localized Cell Death and Extracellular DNA Production in Streptococcus mutans Biofilms
- PMID: 32948520
- PMCID: PMC7657630
- DOI: 10.1128/AEM.02080-20
Competence-Stimulating-Peptide-Dependent Localized Cell Death and Extracellular DNA Production in Streptococcus mutans Biofilms
Abstract
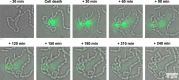
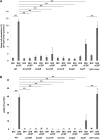
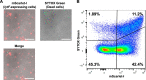
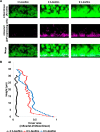
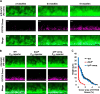
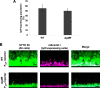
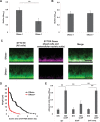
Extracellular DNA (eDNA) is a biofilm component that contributes to the formation and structural stability of biofilms. Streptococcus mutans, a major cariogenic bacterium, induces eDNA-dependent biofilm formation under specific conditions. Since cell death can result in the release and accumulation of DNA, the dead cells in biofilms are a source of eDNA. However, it remains unknown how eDNA is released from dead cells and is localized within S. mutans biofilms. We focused on cell death induced by the extracellular signaling peptide called competence-stimulating peptide (CSP). We demonstrate that nucleic acid release into the extracellular environment occurs in a subpopulation of dead cells. eDNA production induced by CSP was highly dependent on the lytF gene, which encodes an autolysin. Although lytF expression was induced bimodally by CSP, lytF-expressing cells further divided into surviving cells and eDNA-producing dead cells. Moreover, we found that lytF-expressing cells were abundant near the bottom of the biofilm, even when all cells in the biofilm received the CSP signal. Dead cells and eDNA were also abundantly present near the bottom of the biofilm. The number of lytF-expressing cells in biofilms was significantly higher than that in planktonic cultures, which suggests that adhesion to the substratum surface is important for the induction of lytF expression. The deletion of lytF resulted in reduced adherence to a polystyrene surface. These results suggest that lytF expression and eDNA production induced near the bottom of the biofilm contribute to a firmly attached and structurally stable biofilm.IMPORTANCE Bacterial communities encased by self-produced extracellular polymeric substances (EPSs), known as biofilms, have a wide influence on human health and environmental problems. The importance of biofilm research has increased, as biofilms are the preferred bacterial lifestyle in nature. Furthermore, in recent years it has been noted that the contribution of phenotypic heterogeneity within biofilms requires analysis at the single-cell or subpopulation level to understand bacterial life strategies. In Streptococcus mutans, a cariogenic bacterium, extracellular DNA (eDNA) contributes to biofilm formation. However, it remains unclear how and where the cells produce eDNA within the biofilm. We focused on LytF, an autolysin that is induced by extracellular peptide signals. We used single-cell level imaging techniques to analyze lytF expression in the biofilm population. Here, we show that S. mutans generates eDNA by inducing lytF expression near the bottom of the biofilm, thereby enhancing biofilm adhesion and structural stability.
Keywords: Streptococcus mutans; biofilms; cell death; cell-to-cell communication; extracellular DNA.
Copyright © 2020 American Society for Microbiology.
Figures
Similar articles
-
Identification of a Novel Gene Involved in Cell-to-cell Communication-induced Cell Death and eDNA Production in Streptococcus mutans.Microbes Environ. 2023;38(2):ME22085. doi: 10.1264/jsme2.ME22085. Microbes Environ. 2023. PMID: 37302844 Free PMC article.
-
Murein Hydrolase LytF of Streptococcus sanguinis and the Ecological Consequences of Competence Development.Appl Environ Microbiol. 2017 Dec 1;83(24):e01709-17. doi: 10.1128/AEM.01709-17. Print 2017 Dec 15. Appl Environ Microbiol. 2017. PMID: 28986373 Free PMC article.
-
AtlA Mediates Extracellular DNA Release, Which Contributes to Streptococcus mutans Biofilm Formation in an Experimental Rat Model of Infective Endocarditis.Infect Immun. 2017 Aug 18;85(9):e00252-17. doi: 10.1128/IAI.00252-17. Print 2017 Sep. Infect Immun. 2017. PMID: 28674029 Free PMC article.
-
Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms.Front Cell Infect Microbiol. 2015 Feb 13;5:10. doi: 10.3389/fcimb.2015.00010. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 25763359 Free PMC article. Review.
-
Unraveling the multifaceted role of extracellular DNA (eDNA) of biofilm in bacterial physiology, biofilm formation, and matrixome architecture.Crit Rev Biochem Mol Biol. 2025 Feb-Jun;60(1-3):1-32. doi: 10.1080/10409238.2025.2497270. Epub 2025 May 5. Crit Rev Biochem Mol Biol. 2025. PMID: 40322923 Review.
Cited by
-
Dental biofilms contain DNase I-resistant Z-DNA and G-quadruplexes but alternative DNase overcomes this resistance.NPJ Biofilms Microbiomes. 2025 May 19;11(1):80. doi: 10.1038/s41522-025-00694-x. NPJ Biofilms Microbiomes. 2025. PMID: 40389511 Free PMC article.
-
Short-chain fatty acids inhibit the biofilm formation of Streptococcus gordonii through negative regulation of competence-stimulating peptide signaling pathway.J Microbiol. 2021 Dec;59(12):1142-1149. doi: 10.1007/s12275-021-1576-8. Epub 2021 Dec 4. J Microbiol. 2021. PMID: 34865199
-
Effects of lipid emulsions on the formation of Escherichia coli-Candida albicans mixed-species biofilms on PVC.Sci Rep. 2021 Aug 19;11(1):16929. doi: 10.1038/s41598-021-96385-6. Sci Rep. 2021. PMID: 34413406 Free PMC article.
-
Cell differentiation, aging, and death in spatially organized yeast communities: mechanisms and consequences.Cell Death Differ. 2025 Mar 29. doi: 10.1038/s41418-025-01485-9. Online ahead of print. Cell Death Differ. 2025. PMID: 40158069 Review.
-
Identification of a Novel Gene Involved in Cell-to-cell Communication-induced Cell Death and eDNA Production in Streptococcus mutans.Microbes Environ. 2023;38(2):ME22085. doi: 10.1264/jsme2.ME22085. Microbes Environ. 2023. PMID: 37302844 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

