Whole-genome analysis of noncoding genetic variations identifies multiscale regulatory element perturbations associated with Hirschsprung disease
- PMID: 32948616
- PMCID: PMC7605255
- DOI: 10.1101/gr.264473.120
Whole-genome analysis of noncoding genetic variations identifies multiscale regulatory element perturbations associated with Hirschsprung disease
Abstract
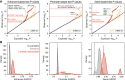
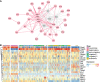
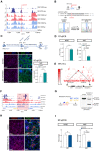
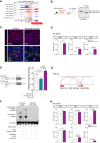
It is widely recognized that noncoding genetic variants play important roles in many human diseases, but there are multiple challenges that hinder the identification of functional disease-associated noncoding variants. The number of noncoding variants can be many times that of coding variants; many of them are not functional but in linkage disequilibrium with the functional ones; different variants can have epistatic effects; different variants can affect the same genes or pathways in different individuals; and some variants are related to each other not by affecting the same gene but by affecting the binding of the same upstream regulator. To overcome these difficulties, we propose a novel analysis framework that considers convergent impacts of different genetic variants on protein binding, which provides multiscale information about disease-associated perturbations of regulatory elements, genes, and pathways. Applying it to our whole-genome sequencing data of 918 short-segment Hirschsprung disease patients and matched controls, we identify various novel genes not detected by standard single-variant and region-based tests, functionally centering on neural crest migration and development. Our framework also identifies upstream regulators whose binding is influenced by the noncoding variants. Using human neural crest cells, we confirm cell stage-specific regulatory roles of three top novel regulatory elements on our list, respectively in the RET, RASGEF1A, and PIK3C2B loci. In the PIK3C2B regulatory element, we further show that a noncoding variant found only in the patients affects the binding of the gliogenesis regulator NFIA, with a corresponding up-regulation of multiple genes in the same topologically associating domain.
© 2020 Fu et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


Similar articles
-
Identification of Genes Associated With Hirschsprung Disease, Based on Whole-Genome Sequence Analysis, and Potential Effects on Enteric Nervous System Development.Gastroenterology. 2018 Dec;155(6):1908-1922.e5. doi: 10.1053/j.gastro.2018.09.012. Epub 2018 Sep 12. Gastroenterology. 2018. PMID: 30217742
-
Variants in RET associated with Hirschsprung's disease affect binding of transcription factors and gene expression.Gastroenterology. 2011 Feb;140(2):572-582.e2. doi: 10.1053/j.gastro.2010.10.044. Epub 2010 Oct 25. Gastroenterology. 2011. PMID: 20977903
-
Whole genome sequencing reveals epistasis effects within RET for Hirschsprung disease.Sci Rep. 2022 Nov 28;12(1):20423. doi: 10.1038/s41598-022-24077-w. Sci Rep. 2022. PMID: 36443333 Free PMC article.
-
Hirschsprung's disease as a model of complex genetic etiology.Histol Histopathol. 2013 Sep;28(9):1117-36. doi: 10.14670/HH-28.1117. Epub 2013 Apr 19. Histol Histopathol. 2013. PMID: 23605783 Review.
-
Hirschsprung disease: Insights on genes, penetrance, and prenatal diagnosis.Neurogastroenterol Motil. 2019 Nov;31(11):e13732. doi: 10.1111/nmo.13732. Neurogastroenterol Motil. 2019. PMID: 31609069 Review.
Cited by
-
Single-cell RNA sequencing in Hirschsprung's disease tissues reveals lack of neuronal differentiation in the aganglionic colon segment.bioRxiv [Preprint]. 2025 Jul 4:2025.07.01.662516. doi: 10.1101/2025.07.01.662516. bioRxiv. 2025. PMID: 40631286 Free PMC article. Preprint.
-
Human Pluripotent Stem Cell-Based Models for Hirschsprung Disease: From 2-D Cell to 3-D Organoid Model.Cells. 2022 Oct 29;11(21):3428. doi: 10.3390/cells11213428. Cells. 2022. PMID: 36359824 Free PMC article. Review.
-
A multi-enhancer RET regulatory code is disrupted in Hirschsprung disease.Genome Res. 2021 Dec;31(12):2199-2208. doi: 10.1101/gr.275667.121. Epub 2021 Nov 15. Genome Res. 2021. PMID: 34782358 Free PMC article.
-
Adult patients with allied disorders of Hirschsprung's disease in emergency department: An 11-year retrospective study.World J Gastrointest Surg. 2022 Jul 27;14(7):656-669. doi: 10.4240/wjgs.v14.i7.656. World J Gastrointest Surg. 2022. PMID: 36158276 Free PMC article.
-
Association between gut microbiota and Hirschsprung disease: a bidirectional two-sample Mendelian randomization study.Front Microbiol. 2024 Mar 7;15:1366181. doi: 10.3389/fmicb.2024.1366181. eCollection 2024. Front Microbiol. 2024. PMID: 38516012 Free PMC article.
References
-
- Alves MM, Sribudiani Y, Brouwer RWW, Amiel J, Antiñolo G, Borrego S, Ceccherini I, Chakravarti A, Fernández RM, Garcia-Barcelo MM, et al. 2013. Contribution of rare and common variants determine complex diseases—Hirschsprung disease as a model. Dev Biol 382: 320–329. 10.1016/j.ydbio.2013.05.019 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
