Dominant mutations in ITPR3 cause Charcot-Marie-Tooth disease
- PMID: 32949214
- PMCID: PMC7545616
- DOI: 10.1002/acn3.51190
Dominant mutations in ITPR3 cause Charcot-Marie-Tooth disease
Abstract
Objective: ITPR3, encoding inositol 1,4,5-trisphosphate receptor type 3, was previously reported as a potential candidate disease gene for Charcot-Marie-Tooth neuropathy. Here, we present genetic and functional evidence that ITPR3 is a Charcot-Marie-Tooth disease gene.
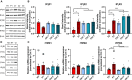
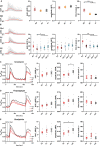
Methods: Whole-exome sequencing of four affected individuals in an autosomal dominant family and one individual who was the only affected individual in his family was used to identify disease-causing variants. Skin fibroblasts from two individuals of the autosomal dominant family were analyzed functionally by western blotting, quantitative reverse transcription PCR, and Ca2+ imaging.
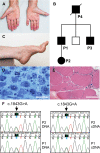
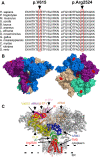
Results: Affected individuals in the autosomal dominant family had onset of symmetrical neuropathy with demyelinating and secondary axonal features at around age 30, showing signs of gradual progression with severe distal leg weakness and hand involvement in the proband at age 64. Exome sequencing identified a heterozygous ITPR3 p.Val615Met variant segregating with the disease. The individual who was the only affected in his family had disease onset at age 4 with demyelinating neuropathy. His condition was progressive, leading to severe muscle atrophy below knees and atrophy of proximal leg and hand muscles by age 16. Trio exome sequencing identified a de novo ITPR3 variant p.Arg2524Cys. Altered Ca2+ -transients in p.Val615Met patient fibroblasts suggested that the variant has a dominant-negative effect on inositol 1,4,5-trisphosphate receptor type 3 function.
Interpretation: Together with two previously identified variants, our report adds further evidence that ITPR3 is a disease-causing gene for CMT and indicates altered Ca2+ homeostasis in disease pathogenesis.
© 2020 The Authors. Annals of Clinical and Translational Neurology published by Wiley Periodicals LLC on behalf of American Neurological Association.
Conflict of interest statement
The authors report no competing interests.
Figures
References
-
- Laura M, Pipis M, Rossor AM, Reilly MM. Charcot‐marie‐tooth disease and related disorders: An evolving landscape. Curr Opin Neurol 2019;32:641–650. - PubMed
-
- Pareyson D, Saveri P, Pisciotta C. New developments in charcot‐marie‐tooth neuropathy and related diseases. Curr Opin Neurol 2017;30:471–480. - PubMed
-
- Schabhuttl M, Wieland T, Senderek J, et al. Whole‐exome sequencing in patients with inherited neuropathies: outcome and challenges. J Neurol 2014;261:970–982. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

