Long-read whole-genome sequencing for the genetic diagnosis of dystrophinopathies
- PMID: 32951359
- PMCID: PMC7545597
- DOI: 10.1002/acn3.51201
Long-read whole-genome sequencing for the genetic diagnosis of dystrophinopathies
Abstract
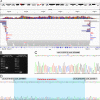
The precise genetic diagnosis of dystrophinopathies can be challenging, largely due to rare deep intronic variants and more complex structural variants (SVs). We report on the genetic characterization of a dystrophinopathy patient. He remained without a genetic diagnosis after routine genetic testing, dystrophin protein and mRNA analysis, and short- and long-read whole DMD gene sequencing. We finally identified a novel complex SV in DMD via long-read whole-genome sequencing. The variant consists of a large-scale (~1Mb) inversion/deletion-insertion rearrangement mediated by LINE-1s. Our study shows that long-read whole-genome sequencing can serve as a clinical diagnostic tool for genetically unsolved dystrophinopathies.
© 2020 The Authors. Annals of Clinical and Translational Neurology published by Wiley Periodicals LLC on behalf of American Neurological Association.
Conflict of interest statement
None.
Figures

References
-
- Aartsma‐Rus A, Van Deutekom JCT, Fokkema IF, et al. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading‐frame rule. Muscle Nerve 2006;34(2):135–144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

