Polymyxin B combinations with FDA-approved non-antibiotic phenothiazine drugs targeting multi-drug resistance of Gram-negative pathogens
- PMID: 32952938
- PMCID: PMC7481501
- DOI: 10.1016/j.csbj.2020.08.008
Polymyxin B combinations with FDA-approved non-antibiotic phenothiazine drugs targeting multi-drug resistance of Gram-negative pathogens
Abstract
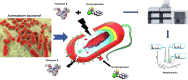
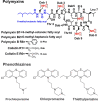
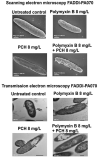
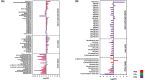
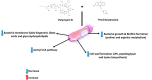
The status quo for combating uprising antibacterial resistance is to employ synergistic combinations of antibiotics. Nevertheless, the currently available combination therapies are fast becoming untenable. Combining antibiotics with various FDA-approved non-antibiotic drugs has emerged as a novel strategy against otherwise untreatable multi-drug resistant (MDR) pathogens. The apex of this study was to investigate the mechanisms of antibacterial synergy of the combination of polymyxin B with the phenothiazines against the MDR Gram-negative pathogens Acinetobacter baumannii, Klebsiella pneumoniae and Pseudomonas aeruginosa. The synergistic antibacterial effects were tested using checkerboard and static time-kill assays. Electron microscopy (EM) and untargeted metabolomics were used to ascertain the mechanism(s) of the antibacterial synergy. The combination of polymyxin B and the phenothiazines showed synergistic antibacterial activity in checkerboard and static time-kill assays at clinically relevant concentrations against both polymyxin-susceptible and polymyxin-resistant isolates. EM revealed that the polymyxin B-prochlorperazine combination resulted in greater damage to the bacterial cell compared to each drug monotherapy. In metabolomics, at 0.5 h, polymyxin B monotherapy and the combination (to a greatest extent) disorganised the bacterial cell envelope as manifested by a major perturbation in bacterial membrane lipids (glycerophospholipids and fatty acids), peptidoglycan and lipopolysaccharide (LPS) biosynthesis. At the late time exposure (4 h), the aforementioned effects (except LPS biosynthesis) perpetuated mainly with the combination therapy, indicating the disorganising bacterial membrane biogenesis is potentially behind the mechanisms of antibacterial synergy. In conclusion, the study highlights the potential usefulness of the combination of polymyxin B with phenothiazines for the treatment of polymyxin-resistant Gram-negative infections (e.g. CNS infections).
Keywords: Antimicrobial peptides; Antimicrobial resistance; Gram-negative; MDR; Metabolomics; Phenothiazines.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Mechanisms Underlying Synergistic Killing of Polymyxin B in Combination with Cannabidiol against Acinetobacter baumannii: A Metabolomic Study.Pharmaceutics. 2022 Apr 3;14(4):786. doi: 10.3390/pharmaceutics14040786. Pharmaceutics. 2022. PMID: 35456620 Free PMC article.
-
Effective Strategy Targeting Polymyxin-Resistant Gram-Negative Pathogens: Polymyxin B in Combination with the Selective Serotonin Reuptake Inhibitor Sertraline.ACS Infect Dis. 2020 Jun 12;6(6):1436-1450. doi: 10.1021/acsinfecdis.0c00108. Epub 2020 May 25. ACS Infect Dis. 2020. PMID: 32427476
-
From Breast Cancer to Antimicrobial: Combating Extremely Resistant Gram-Negative "Superbugs" Using Novel Combinations of Polymyxin B with Selective Estrogen Receptor Modulators.Microb Drug Resist. 2017 Jul;23(5):640-650. doi: 10.1089/mdr.2016.0196. Epub 2016 Dec 9. Microb Drug Resist. 2017. PMID: 27935770
-
Antibiotic-non-antibiotic combinations for combating extremely drug-resistant Gram-negative 'superbugs'.Essays Biochem. 2017 Mar 3;61(1):115-125. doi: 10.1042/EBC20160058. Print 2017 Feb 28. Essays Biochem. 2017. PMID: 28258235 Review.
-
Polymyxin combination therapy for multidrug-resistant, extensively-drug resistant, and difficult-to-treat drug-resistant gram-negative infections: is it superior to polymyxin monotherapy?Expert Rev Anti Infect Ther. 2023 Apr;21(4):387-429. doi: 10.1080/14787210.2023.2184346. Epub 2023 Mar 8. Expert Rev Anti Infect Ther. 2023. PMID: 36820511 Review.
Cited by
-
Integrated Transcriptomic and Metabolomic Mapping Reveals the Mechanism of Action of Ceftazidime/Avibactam against Pan-Drug-Resistant Klebsiella pneumoniae.ACS Infect Dis. 2023 Dec 8;9(12):2409-2422. doi: 10.1021/acsinfecdis.3c00264. Epub 2023 Oct 25. ACS Infect Dis. 2023. PMID: 37878861 Free PMC article.
-
Armored polymyxin B: a nanosystem for combating multidrug-resistant Gram-negative bacilli.RSC Adv. 2024 Dec 17;14(53):39700-39707. doi: 10.1039/d4ra07577c. eCollection 2024 Dec 10. RSC Adv. 2024. PMID: 39691227 Free PMC article.
-
Alkyl deoxyglycoside-polymyxin combinations against critical priority carbapenem-resistant gram-negative bacteria.Sci Rep. 2024 Jan 26;14(1):2219. doi: 10.1038/s41598-024-51428-6. Sci Rep. 2024. PMID: 38278870 Free PMC article.
-
Unique mechanistic insights into pathways associated with the synergistic activity of polymyxin B and caspofungin against multidrug-resistant Klebsiella pneumoniae.Comput Struct Biotechnol J. 2022 Feb 25;20:1077-1087. doi: 10.1016/j.csbj.2022.02.021. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35284046 Free PMC article.
-
Mechanisms Underlying Synergistic Killing of Polymyxin B in Combination with Cannabidiol against Acinetobacter baumannii: A Metabolomic Study.Pharmaceutics. 2022 Apr 3;14(4):786. doi: 10.3390/pharmaceutics14040786. Pharmaceutics. 2022. PMID: 35456620 Free PMC article.
References
-
- WHO. Antimicrobial resistance: global report on surveillance 2014. 2014.
-
- Organization W.H. WHO; Geneva: 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics.
-
- Boucher H.W., Talbot G.H., Bradley J.S., Edwards J.E., Gilbert D., Rice L.B. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. - PubMed
-
- Rice L.B. The University of Chicago Press; 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. - PubMed
-
- Breidenstein E.B., de la Fuente-Nunez C., Hancock R.E. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19:419–426. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
