PI3K inhibitors: review and new strategies
- PMID: 32953006
- PMCID: PMC7472334
- DOI: 10.1039/d0sc01676d
PI3K inhibitors: review and new strategies
Abstract
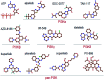
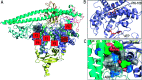
The search is on for effective specific inhibitors for PI3Kα mutants. PI3Kα, a critical lipid kinase, has two subunits, catalytic and inhibitory. PIK3CA, the gene that encodes the p110α catalytic subunit is a highly mutated protein in cancer. Dysregulation of PI3Kα signalling is commonly associated with tumorigenesis and drug resistance. Despite its vast importance, only recently the FDA approved the first drug (alpelisib by Novartis) for breast cancer. A second (GDC0077), classified as PI3Kα isoform-specific, is undergoing clinical trials. Not surprisingly, these ATP-competitive drugs commonly elicit severe concentration-dependent side effects. Here we briefly review PI3Kα mutations, focus on PI3K drug repertoire and propose new, to-date unexplored PI3Kα therapeutic strategies. These include (1) an allosteric and orthosteric inhibitor combination and (2) taking advantage of allosteric rescue mutations to guide drug discovery.
This journal is © The Royal Society of Chemistry 2020.
Figures


Similar articles
-
Allosteric PI3Kα Inhibition Overcomes On-target Resistance to Orthosteric Inhibitors Mediated by Secondary PIK3CA Mutations.Cancer Discov. 2024 Feb 8;14(2):227-239. doi: 10.1158/2159-8290.CD-23-0704. Cancer Discov. 2024. PMID: 37916958 Free PMC article.
-
Development of PI3Kα inhibitors for tumor therapy.J Biomol Struct Dyn. 2023 Oct-Nov;41(17):8587-8604. doi: 10.1080/07391102.2022.2132293. Epub 2022 Oct 11. J Biomol Struct Dyn. 2023. PMID: 36221910 Review.
-
Comparative molecular dynamics analyses on PIK3CA hotspot mutations with PI3Kα specific inhibitors and ATP.Comput Biol Chem. 2022 Aug;99:107726. doi: 10.1016/j.compbiolchem.2022.107726. Epub 2022 Jul 8. Comput Biol Chem. 2022. PMID: 35842959
-
Oncogenic activation of PI K3 CA in cancers: Emerging targeted therapies in precision oncology.Genes Dis. 2024 Sep 10;12(2):101430. doi: 10.1016/j.gendis.2024.101430. eCollection 2025 Mar. Genes Dis. 2024. PMID: 39717717 Free PMC article. Review.
-
Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3Kα Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.Cancer Discov. 2024 Feb 8;14(2):240-257. doi: 10.1158/2159-8290.CD-23-0944. Cancer Discov. 2024. PMID: 37916956 Free PMC article.
Cited by
-
The emerging role of PI3K inhibitors for solid tumour treatment and beyond.Br J Cancer. 2023 Jun;128(12):2150-2162. doi: 10.1038/s41416-023-02221-1. Epub 2023 Mar 13. Br J Cancer. 2023. PMID: 36914722 Free PMC article. Review.
-
Heparin/Collagen-REDV Modification of Expanded Polytetrafluoroethylene Improves Regional Anti-thrombosis and Reduces Foreign Body Reactions in Local Tissues.Front Bioeng Biotechnol. 2022 Aug 4;10:916931. doi: 10.3389/fbioe.2022.916931. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35992343 Free PMC article.
-
Discovery of cryptic allosteric sites using reversed allosteric communication by a combined computational and experimental strategy.Chem Sci. 2020 Nov 2;12(1):464-476. doi: 10.1039/d0sc05131d. Chem Sci. 2020. PMID: 34163609 Free PMC article.
-
Liquid biopsy: Cell-free DNA based analysis in breast cancer.J Liq Biopsy. 2023 Jul 27;1:100002. doi: 10.1016/j.jlb.2023.100002. eCollection 2023 Sep. J Liq Biopsy. 2023. PMID: 40027284 Free PMC article. Review.
-
Mechanism of activation and the rewired network: New drug design concepts.Med Res Rev. 2022 Mar;42(2):770-799. doi: 10.1002/med.21863. Epub 2021 Oct 25. Med Res Rev. 2022. PMID: 34693559 Free PMC article. Review.
References
-
- Carrera A. C., Anderson R. J. Cell Sci. 2019;132:jcs228395. - PubMed
-
- S. I. C. database, http://www.sanger.ac.uk/genetics/CGP/cosmic/.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

