Genetic and microenvironmental differences in non-smoking lung adenocarcinoma patients compared with smoking patients
- PMID: 32953513
- PMCID: PMC7481643
- DOI: 10.21037/tlcr-20-276
Genetic and microenvironmental differences in non-smoking lung adenocarcinoma patients compared with smoking patients
Abstract
Background: Non-smoking-related lung adenocarcinoma (LUAD) has its own characteristics. Genetic and microenvironmental differences in smoking and non-smoking LUAD patients were analyzed to elucidate the oncogenesis of non-smoking-related LUAD, which will improve our understanding of the underlying molecular mechanism and be of clinical use in the future.
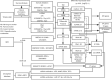
Methods: The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) databases were used for clinical and genomic information. Various bioinformatics tools were used to analyze differences in somatic mutations, RNA and microRNA (miRNA) expression, immune infiltration, and stemness indices. GO, KEGG, and GSVA analyses were performed with R. A merged protein-protein interaction (PPI) network was constructed and analyzed. A miRNA-differentially expressed gene network was constructed with miRNet. qRT-PCR was used for validation of 4 most significantly differently expressed genes and 2 miRNAs in tumor samples obtained from 20 pairs of non-smoking and smoking patients.
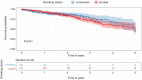
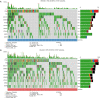
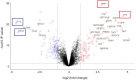
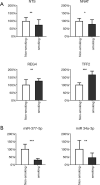
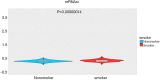
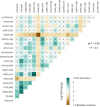
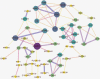
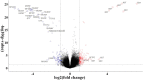
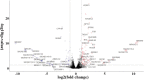
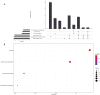
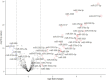
Results: Five hundred and one patients with LUAD were obtained, including 210 in the non-smoking group and 292 in the smoking group. A total of 174 significantly altered somatic mutations were detected, including mutations in tumor protein p53 and epidermal growth factor receptor, which were downregulated in non-smoking-related LUAD. At the RNA level, 231 significantly differentially expressed genes were obtained; 124 were upregulated and 107 downregulated in the non-smoking group. GSVA analysis revealed 42 significant pathways. Other functional and enrichment analyses of somatic mutations and RNA expression levels revealed that these genes were significantly enriched in receptor activity regulation and receptor binding. Differences in microenvironments including immune infiltration (e.g., CD8+ T cells and resting mast cells) and stemness indices were also found between groups. A 79-pair interaction was found between differentially expressed genes and miRNAs, of which miR-335-5p and miR-34a-5p were located in the center. Twenty-one genes, including vitronectin, neurotensin, and neuronatin, were differentially expressed in both non-smoking LUAD patients and DMSO-treated A549 cells. And the different expression of neurotensin, neuronatin, trefoil factor family2, regenerating family member 4, miR-377-5p, miR-34a were verified with the same tendency in our own samples.
Conclusions: Non-smoking LUAD patients, compared to smokers, have different characteristics in terms of somatic mutation, gene, and miRNA expression and the microenvironment, indicating a diverse mechanism of oncogenesis.
Keywords: Lung adenocarcinoma; genome; microenvironment; non-smoking.
2020 Translational Lung Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-276). The authors have no conflicts of interest to declare.
Figures




Similar articles
-
Differences in genetics and microenvironment of lung adenocarcinoma patients with or without TP53 mutation.BMC Pulm Med. 2021 Oct 11;21(1):316. doi: 10.1186/s12890-021-01671-8. BMC Pulm Med. 2021. PMID: 34635074 Free PMC article.
-
Clinical value of microRNA-198-5p downregulation in lung adenocarcinoma and its potential pathways.Oncol Lett. 2019 Sep;18(3):2939-2954. doi: 10.3892/ol.2019.10610. Epub 2019 Jul 12. Oncol Lett. 2019. PMID: 31402959 Free PMC article.
-
Integrated analysis of patients with KEAP1/NFE2L2/CUL3 mutations in lung adenocarcinomas.Cancer Med. 2021 Dec;10(23):8673-8692. doi: 10.1002/cam4.4338. Epub 2021 Oct 6. Cancer Med. 2021. PMID: 34617407 Free PMC article.
-
Expression, Clinical Significance, Immune Infiltration, and Regulation Network of miR-3940-5p in Lung Adenocarcinoma Based on Bioinformatic Analysis and Experimental Validation.Int J Gen Med. 2022 Aug 6;15:6451-6464. doi: 10.2147/IJGM.S375761. eCollection 2022. Int J Gen Med. 2022. PMID: 35966511 Free PMC article.
-
Identification key genes, key miRNAs and key transcription factors of lung adenocarcinoma.J Thorac Dis. 2020 May;12(5):1917-1933. doi: 10.21037/jtd-19-4168. J Thorac Dis. 2020. PMID: 32642095 Free PMC article.
Cited by
-
Unveiling the mechanisms and challenges of cancer drug resistance.Cell Commun Signal. 2024 Feb 12;22(1):109. doi: 10.1186/s12964-023-01302-1. Cell Commun Signal. 2024. PMID: 38347575 Free PMC article. Review.
-
Tumour and microenvironment crosstalk in NSCLC progression and response to therapy.Nat Rev Clin Oncol. 2025 Jul;22(7):463-482. doi: 10.1038/s41571-025-01021-1. Epub 2025 May 16. Nat Rev Clin Oncol. 2025. PMID: 40379986 Free PMC article. Review.
-
Anti-tumor effects of miR-34a by regulating immune cells in the tumor microenvironment.Cancer Med. 2023 May;12(10):11602-11610. doi: 10.1002/cam4.5826. Epub 2023 Mar 23. Cancer Med. 2023. PMID: 36951490 Free PMC article. Review.
-
MOCSS: Multi-omics data clustering and cancer subtyping via shared and specific representation learning.iScience. 2023 Jul 13;26(8):107378. doi: 10.1016/j.isci.2023.107378. eCollection 2023 Aug 18. iScience. 2023. PMID: 37559907 Free PMC article.
-
Survival benefit of surgery vs radiotherapy alone to patients with stage IA lung adenocarcinoma: a propensity score-matched analysis.Eur J Med Res. 2025 Mar 15;30(1):173. doi: 10.1186/s40001-025-02436-3. Eur J Med Res. 2025. PMID: 40089771 Free PMC article.
References
-
- WHO report on the global tobacco epidemic 2019. Available online: https://apps.who.int/iris/bitstream/handle/10665/326043/9789241516204-en..., accessed 12 December 2019: Geneva: World Health Organization 2019.
-
- WHO highlights huge scale of tobacco-related lung disease deaths. In: World No Tobacco Day 2019: Don’t let tobacco take your breath away. Geneva: World Health Organization. Available online: https://www.who.int/news-room/detail/29-05-2019-who-highlights-huge-scal.... Accessed 12/12 2019.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
