High-resolution epitope mapping of anti-Hu and anti-Yo autoimmunity by programmable phage display
- PMID: 32954318
- PMCID: PMC7425417
- DOI: 10.1093/braincomms/fcaa059
High-resolution epitope mapping of anti-Hu and anti-Yo autoimmunity by programmable phage display
Abstract
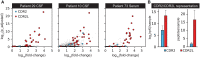
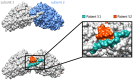
Paraneoplastic neurological disorders are immune-mediated diseases understood to manifest as part of a misdirected anti-tumor immune response. Paraneoplastic neurological disorder-associated autoantibodies can assist with diagnosis and enhance our understanding of tumor-associated immune processes. We designed a comprehensive library of 49-amino-acid overlapping peptides spanning the entire human proteome, including all splicing isoforms and computationally predicted coding regions. Using this library, we optimized a phage immunoprecipitation and sequencing protocol with multiple rounds of enrichment to create high-resolution epitope profiles in serum and cerebrospinal fluid (CSF) samples from patients suffering from two common paraneoplastic neurological disorders, the anti-Yo (n = 36 patients) and anti-Hu (n = 44 patients) syndromes. All (100%) anti-Yo patient samples yielded enrichment of peptides from the canonical anti-Yo (CDR2 and CDR2L) antigens, while 38% of anti-Hu patients enriched peptides deriving from the nELAVL (neuronal embryonic lethal abnormal vision like) family of proteins, the anti-Hu autoantigenic target. Among the anti-Hu patient samples that were positive for nELAVL, we noted a restricted region of immunoreactivity. To achieve single amino acid resolution, we designed a novel deep mutational scanning phage library encoding all possible single-point mutants targeting the reactive nELAVL region. This analysis revealed a distinct preference for the degenerate motif, RLDxLL, shared by ELAVL2, 3 and 4. Lastly, phage immunoprecipitation sequencing identified several known autoantigens in these same patient samples, including peptides deriving from the cancer-associated antigens ZIC and SOX families of transcription factors. Overall, this optimized phage immunoprecipitation sequencing library and protocol yielded the high-resolution epitope mapping of the autoantigens targeted in anti-Yo and anti-Hu encephalitis patients to date. The results presented here further demonstrate the utility and high-resolution capability of phage immunoprecipitation sequencing for both basic science and clinical applications and for better understanding the antigenic targets and triggers of paraneoplastic neurological disorders.
Keywords: anti-Hu; anti-Yo; autoimmunity; paraneoplastic; phage display.
© The Author(s) (2020). Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures






References
-
- Beghetto E, Gargano N.. Antigen discovery using whole-genome phage display libraries. Methods Mol Biol 2013; 1061: 79–95. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 1995; 57: 289–300.
-
- Burritt JB, Quinn MT, Jutila MA, Bond CW, Jesaitis AJ.. Topological mapping of neutrophil cytochrome b epitopes with phage-display libraries. J Biol Chem 1995; 270: 16974–80. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
