A regulatory region on RIPK2 is required for XIAP binding and NOD signaling activity
- PMID: 32954645
- PMCID: PMC7645253
- DOI: 10.15252/embr.202050400
A regulatory region on RIPK2 is required for XIAP binding and NOD signaling activity
Abstract
Signaling via the intracellular pathogen receptors nucleotide-binding oligomerization domain-containing proteins NOD1 and NOD2 requires receptor interacting kinase 2 (RIPK2), an adaptor kinase that can be targeted for the treatment of various inflammatory diseases. However, the molecular mechanisms of how RIPK2 contributes to NOD signaling are not completely understood. We generated FLAG-tagged RIPK2 knock-in mice using CRISPR/Cas9 technology to study NOD signaling mechanisms at the endogenous level. Using cells from these mice, we were able to generate a detailed map of post-translational modifications on RIPK2. Similar to other reports, we did not detect ubiquitination of RIPK2 lysine 209 during NOD2 signaling. However, using site-directed mutagenesis we identified a new regulatory region on RIPK2, which dictates the crucial interaction with the E3 ligase XIAP and downstream signaling outcomes.
Keywords: XIAP; NOD signaling; RIPK2; inflammation; ubiquitin.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Tissue distribution of RIPK2 determined by anti‐FLAG immunoprecipitation. Organ homogenates from WT (wild‐type) and KI (homozygous FLAG‐RIPK2) mice were subjected to anti‐FLAG immunoprecipitation and immunoblotting.
Inflammatory signaling in wild‐type (WT/WT), RIPK2 CRISPR KO (KO/KO), and FLAG‐RIPK2 heterozygous (WT/KI) and homozygous (KI/KI) BMDMs. BMDMs were primed with IFNγ, stimulated with MDP for indicated times, and analyzed by immunoblotting.
Cytokine production of RIPK2 CRISPR KO (KO), wild‐type (WT), and FLAG‐RIPK2 heterozygous (WT/KI) and homozygous (KI/KI) BMDMs in response to MDP. BMDMs were left untreated or treated with IFNγ alone or IFNγ and MDP overnight and cytokines were measured by ELISA. N = 5–8 mice. Shown is average ± SEM. ns P > 0.05; **P ≤ 0.01; ****P ≤ 0.0001; two‐way ANOVA.
Serum cytokines in RIPK2 CRISPR KO (KO), wild‐type (WT), and FLAG‐RIPK2 heterozygous (WT/KI) or homozygous (KI/KI) mice after i.p. MDP administration. Mice were injected i.p. with PBS or MDP, sacrificed after 4 h and serum cytokines were measured by ELISA. N = 3–6 mice. Shown is average ± SEM. ns P > 0.05; **P ≤ 0.01; two‐way ANOVA.
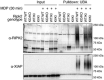

Two‐step enrichment to isolate ubiquitinated RIPK2 from BMDMs. Protein lysates from FLAG‐RIPK2 BMDMs (A) were sequentially subjected to ubiquitin enrichment (UBA, B) and FLAG pulldown (C) prior to protein elution and subsequent mass spectrometry analysis.
Schematic representation of RIPK2 PTMs detected in MDP‐stimulated vs. unstimulated BMDMs. Red: phosphorylation, green: ubiquitination. N = 3 experiments.
DiGly modifications on RIPK2 in L18‐MDP-stimulated vs. L18‐MDP-unstimulated THP‐1 cells determined by diGly proteomics. Shown are P values and log2 differences of modified peptides based on imputed values; t‐test. Score: peptide identification score as determined by MaxQuant.
Sequence conservation of stimulation‐dependent modified serine (red) and lysine (green) residues among mammals. The degree of conservation is indicated by color saturation.

Activation of the NF‐κB pathway by RIPK2 Lysine‐ and phosphosite mutants. RIPK2‐deficient THP‐1 cells reconstituted with wild‐type RIPK2 or RIPK2 mutants were stimulated with L18‐MDP, harvested at indicated time points and activation of the NF‐κB pathway was analyzed by immunoblotting.
IL‐8 production of wild‐type THP‐1 and RIPK2‐deficient THP‐1 cells reconstituted with wild‐type RIPK2 or RIPK2 mutants and stimulated with L18‐MDP was assessed by ELISA. N = 4–8 experiments. Shown is average ± SEM. *P ≤ 0.05; two‐way ANOVA.
RIPK2 ubiquitination determined by UBA pulldown in RIPK2‐deficient cells reconstituted with wild‐type or mutant RIPK2 after stimulation with L18‐MDP.
Detection of K63‐ and M1‐linked ubiquitin chains by UBA pulldown or pulldown with ubiquitin chain type‐specific antibodies in RIPK2‐deficient cells reconstituted with wild‐type or K209R RIPK2 after stimulation with L18‐MDP.

Structural features of the RIPK2 kinase domain (left) and location of K209 within a hydrophobic pocket between helices αEF and αE (right). Shown is chain B of the RIPK2 kinase in complex with ponatinib from PDB:4C8B. The electrostatic interaction potential is shown as a blue to red gradient.
Conservation of amino acids creating a hydrophobic pocket between helices αEF and αE. Degree of conservation among 227 vertebrate species indicated by background color saturation.
Activation of the NF‐κB pathway by RIPK2 pocket mutants. RIPK2‐deficient THP‐1 cells were reconstituted with wild‐type RIPK2 or RIPK2 mutants, stimulated with L18‐MDP, harvested at indicated time points, and analyzed by immunoblotting.
IL‐8 production of RIPK2‐deficient THP‐1 cells reconstituted with wild‐type RIPK2 or RIPK2 mutants and stimulated with L18‐MDP was assessed by ELISA. N = 3–8 experiments. Shown is average ± SEM. ns P > 0.05; *P ≤ 0.05; ***P ≤ 0.001; ****P ≤ 0.0001; two‐way ANOVA.
Ubiquitination of RIPK2 pocket mutants. RIPK2‐deficient THP‐1 cells were reconstituted with wild‐type or mutant RIPK2, left unstimulated or stimulated with L18‐MDP and subjected to UBA pulldown and immunoblotting.

Thermal stability assay using selected RIPK2 mutants. THP‐1 cells expressing RIPK2 were subjected to heat treatment, and non‐denatured fractions were analyzed by immunoblotting.
Binding of RIPK2 to XIAP‐BIR2. Lysates from RIPK2‐deficient THP‐1 cells reconstituted with WT or mutant RIPK2 were subjected to pulldown experiments with recombinantly expressed XIAP‐BIR2 and analyzed by immunoblotting.

References
-
- Aebersold R, Mann M (2003) Mass spectrometry‐based proteomics. Nature 422: 198–207 - PubMed
-
- Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermudez‐Humaran LG, Smirnova N, Berge M, Sulpice T, Lahtinen S et al (2011) Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med 3: 559–572 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1046986/Department of Health | National Health and Medical Research Council (NHMRC)
- 1057888/Department of Health | National Health and Medical Research Council (NHMRC)
- 541901/Department of Health | National Health and Medical Research Council (NHMRC)
- 1058190/Department of Health | National Health and Medical Research Council (NHMRC)
- 9000220, 1162058/Department of Health | National Health and Medical Research Council (NHMRC)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

