Automated SARS-COV-2 RNA extraction from patient nasopharyngeal samples using a modified DNA extraction kit for high throughput testing
- PMID: 32954791
- PMCID: PMC7511039
- DOI: 10.5144/0256-4947.2020.373
Automated SARS-COV-2 RNA extraction from patient nasopharyngeal samples using a modified DNA extraction kit for high throughput testing
Abstract
Background: The pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) has prompted a need for mass testing to identify patients with viral infection. The high demand has created a global bottleneck in testing capacity, which prompted us to modify available resources to extract viral RNA and perform reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) to detect SARS-COV-2.
Objectives: Report on the use of a DNA extraction kit, after modifications, to extract viral RNA that could then be detected using an FDA-approved SARS-COV-2 RT-qPCR assay.
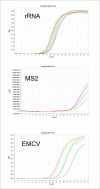
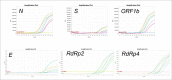
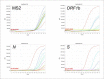
Materials and methods: Initially, automated RNA extraction was performed using a modified DNA kit on samples from control subjects, a bacteriophage, and an RNA virus. We then verified the automated extraction using the modified kit to detect in-lab propagated SARSCOV-2 titrations using an FDA approved commercial kit (S, N, and ORF1b genes) and an in-house primer-probe based assay (E, RdRp2 and RdRp4 genes).
Results: Automated RNA extraction on serial dilutions SARS-COV-2 achieved successful one-step RT-qPCR detection down to 60 copies using the commercial kit assay and less than 30 copies using the in-house primer-probe assay. Moreover, RT-qPCR detection was successful after automated RNA extraction using this modified protocol on 12 patient samples of SARS-COV-2 collected by nasopharyngeal swabs and stored in viral transport media.
Conclusions: We demonstrated the capacity of a modified DNA extraction kit for automated viral RNA extraction and detection using a platform that is suitable for mass testing.
Limitations: Small patient sample size.
Conflict of interest: None.
Figures
Similar articles
-
The Allplex 2019-nCoV (Seegene) assay: which performances are for SARS-CoV-2 infection diagnosis?Eur J Clin Microbiol Infect Dis. 2020 Oct;39(10):1997-2000. doi: 10.1007/s10096-020-03930-8. Epub 2020 May 28. Eur J Clin Microbiol Infect Dis. 2020. PMID: 32462501 Free PMC article.
-
Does bronchoscopy help the diagnosis in COVID-19 infection?Eur Respir J. 2020 Aug 6;56(2):2001619. doi: 10.1183/13993003.01619-2020. Print 2020 Aug. Eur Respir J. 2020. PMID: 32527742 Free PMC article.
-
Rapid random access detection of the novel SARS-coronavirus-2 (SARS-CoV-2, previously 2019-nCoV) using an open access protocol for the Panther Fusion.J Clin Virol. 2020 Apr;125:104305. doi: 10.1016/j.jcv.2020.104305. Epub 2020 Feb 28. J Clin Virol. 2020. PMID: 32143123 Free PMC article.
-
The genetic sequence, origin, and diagnosis of SARS-CoV-2.Eur J Clin Microbiol Infect Dis. 2020 Sep;39(9):1629-1635. doi: 10.1007/s10096-020-03899-4. Epub 2020 Apr 24. Eur J Clin Microbiol Infect Dis. 2020. PMID: 32333222 Free PMC article. Review.
-
The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19.Protein J. 2020 Jun;39(3):198-216. doi: 10.1007/s10930-020-09901-4. Protein J. 2020. PMID: 32447571 Free PMC article. Review.
Cited by
-
Comparative Evaluation of Six SARS-CoV-2 Real-Time RT-PCR Diagnostic Approaches Shows Substantial Genomic Variant-Dependent Intra- and Inter-Test Variability, Poor Interchangeability of Cycle Threshold and Complementary Turn-Around Times.Pathogens. 2022 Apr 12;11(4):462. doi: 10.3390/pathogens11040462. Pathogens. 2022. PMID: 35456137 Free PMC article.
-
Novel miniaturized fluorescence loop-mediated isothermal amplification detection system for rapid on-site virus detection.Front Bioeng Biotechnol. 2022 Aug 24;10:964244. doi: 10.3389/fbioe.2022.964244. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36091427 Free PMC article.
-
An open-source, automated, and cost-effective platform for COVID-19 diagnosis and rapid portable genomic surveillance using nanopore sequencing.Sci Rep. 2023 Nov 21;13(1):20349. doi: 10.1038/s41598-023-47190-w. Sci Rep. 2023. PMID: 37990068 Free PMC article.
-
Navigating the Post-COVID-19 Immunological Era: Understanding Long COVID-19 and Immune Response.Life (Basel). 2023 Oct 26;13(11):2121. doi: 10.3390/life13112121. Life (Basel). 2023. PMID: 38004261 Free PMC article. Review.
-
Effect of gamma irradiation on filtering facepiece respirators and SARS-CoV-2 detection.Sci Rep. 2021 Oct 6;11(1):19888. doi: 10.1038/s41598-021-99414-6. Sci Rep. 2021. PMID: 34615977 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

