A Model Membrane Platform for Reconstituting Mitochondrial Membrane Dynamics
- PMID: 32955498
- PMCID: PMC11153653
- DOI: 10.3791/61620
A Model Membrane Platform for Reconstituting Mitochondrial Membrane Dynamics
Abstract
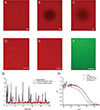
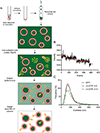
Mitochondrial dynamics is essential for the organelle's diverse functions and cellular responses. The crowded, spatially complex, mitochondrial membrane is a challenging environment to distinguish regulatory factors. Experimental control of protein and lipid components can help answer specific questions of regulation. Yet, quantitative manipulation of these factors is challenging in cellular assays. To investigate the molecular mechanism of mitochondria inner-membrane fusion, we introduced an in vitro reconstitution platform that mimics the lipid environment of the mitochondrial inner-membrane. Here we describe detailed steps for preparing lipid bilayers and reconstituting mitochondrial membrane proteins. The platform allowed analysis of intermediates in mitochondrial inner-membrane fusion, and the kinetics for individual transitions, in a quantitative manner. This protocol describes the fabrication of bilayers with asymmetric lipid composition and describes general considerations for reconstituting transmembrane proteins into a cushioned bilayer. The method may be applied to study other membrane systems.
Figures




References
-
- Sackmann E, Biological membranes architecture and function. In Structure and Dynamics of Membranes. Ed: Lipowsky R, Sackmann E. Elsiever. 1, 1–63 (1995).
-
- Rajendran L, Simons K, Lipid rafts and membrane dynamics. Journal of Cell Science. 118 (Pt 6), 1099–1102 (2005). - PubMed
-
- Schink KO, Tan KW, Stenmark H Phosphoinositides in Control of Membrane Dynamics. Annual Review of Cell and Developmental Biology. 32, 143–171 (2016). - PubMed
-
- Schafer DA Coupling actin dynamics and membrane dynamics during endocytosis. Current Opinion in Cell Biology. 14 (1), 76–81 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
