Coordinate genomic association of transcription factors controlled by an imported quorum sensing peptide in Cryptococcus neoformans
- PMID: 32956370
- PMCID: PMC7537855
- DOI: 10.1371/journal.pgen.1008744
Coordinate genomic association of transcription factors controlled by an imported quorum sensing peptide in Cryptococcus neoformans
Abstract
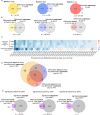
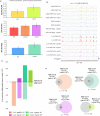
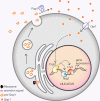
Qsp1 is a secreted quorum sensing peptide required for virulence of the fungal meningitis pathogen Cryptococcus neoformans. Qsp1 functions to control cell wall integrity in vegetatively growing cells and also functions in mating. Rather than acting on a cell surface receptor, Qsp1 is imported to act intracellularly via the predicted oligopeptide transporter Opt1. Here, we identify a transcription factor network as a target of Qsp1. Using whole-genome chromatin immunoprecipitation, we find Qsp1 controls the genomic associations of three transcription factors to genes whose outputs are regulated by Qsp1. One of these transcription factors, Cqs2, is also required for the action of Qsp1 during mating, indicating that it might be a shared proximal target of Qsp1. Consistent with this hypothesis, deletion of CQS2 impacts the binding of other network transcription factors specifically to Qsp1-regulated genes. These genetic and genomic studies illuminate mechanisms by which an imported peptide acts to modulate eukaryotic gene expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Bassler BL, Miller MB. Quorum Sensing In: Rosenberg E, DeLong EF, S Lory, E Stackebrandt, Thompson F, editors. The Prokaryotes: Prokaryotic Communities and Ecophysiology [Internet]. Berlin, Heidelberg: Springer; 2013. [cited 2020 Feb 29]. p. 495–509. Available from: 10.1007/978-3-642-30123-0_60 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

