Cooperativity between the Ribosome-Associated Chaperone Ssb/RAC and the Ubiquitin Ligase Ltn1 in Ubiquitination of Nascent Polypeptides
- PMID: 32957466
- PMCID: PMC7554835
- DOI: 10.3390/ijms21186815
Cooperativity between the Ribosome-Associated Chaperone Ssb/RAC and the Ubiquitin Ligase Ltn1 in Ubiquitination of Nascent Polypeptides
Abstract
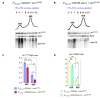
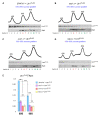
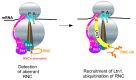
Eukaryotic cells have evolved multiple mechanisms to detect and eliminate aberrant polypeptides. Co-translational protein surveillance systems play an important role in these mechanisms. These systems include ribosome-associated protein quality control (RQC) that detects aberrant nascent chains stalled on ribosomes and promotes their ubiquitination and degradation by the proteasome, and ribosome-associated chaperone Ssb/RAC, which ensures correct nascent chain folding. Despite the known function of RQC and Ssb/ribosome-associated complex (RAC) in monitoring the quality of newly generated polypeptides, whether they cooperate during initial stages of protein synthesis remains unexplored. Here, we provide evidence that Ssb/RAC and the ubiquitin ligase Ltn1, the major component of RQC, display genetic and functional cooperativity. Overexpression of Ltn1 rescues growth suppression of the yeast strain-bearing deletions of SSB genes during proteotoxic stress. Moreover, Ssb/RAC promotes Ltn1-dependent ubiquitination of nascent chains associated with 80S ribosomal particles but not with translating ribosomes. Consistent with this finding, quantitative western blot analysis revealed lower levels of Ltn1 associated with 80S ribosomes and with free 60S ribosomal subunits in the absence of Ssb/RAC. We propose a mechanism in which Ssb/RAC facilitates recruitment of Ltn1 to ribosomes, likely by detecting aberrations in nascent chains and leading to their ubiquitination and degradation.
Keywords: Ssb/RAC triad; r-protein; rRNA; ribosome; ribosome-associated chaperones; ribosome-associated protein quality control (RQC); ribosome-bound nascent chains (RNCs); ubiquitin; ubiquitin ligase Ltn1; ubiquitination of polypeptides bound to a ribosome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Rqc1 and Ltn1 Prevent C-terminal Alanine-Threonine Tail (CAT-tail)-induced Protein Aggregation by Efficient Recruitment of Cdc48 on Stalled 60S Subunits.J Biol Chem. 2016 Jun 3;291(23):12245-53. doi: 10.1074/jbc.M116.722264. Epub 2016 Apr 18. J Biol Chem. 2016. PMID: 27129255 Free PMC article.
-
Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex.Proc Natl Acad Sci U S A. 2014 Nov 11;111(45):15981-6. doi: 10.1073/pnas.1413882111. Epub 2014 Oct 27. Proc Natl Acad Sci U S A. 2014. PMID: 25349383 Free PMC article.
-
Mechanism of nascent chain removal by the ribosome-associated quality control complex.Nat Commun. 2025 Jul 1;16(1):5792. doi: 10.1038/s41467-025-61235-w. Nat Commun. 2025. PMID: 40595698 Free PMC article.
-
Ribosome-associated quality-control mechanisms from bacteria to humans.Mol Cell. 2022 Apr 21;82(8):1451-1466. doi: 10.1016/j.molcel.2022.03.038. Mol Cell. 2022. PMID: 35452614 Free PMC article. Review.
-
Argonaute-dependent ribosome-associated protein quality control.Trends Cell Biol. 2023 Mar;33(3):260-272. doi: 10.1016/j.tcb.2022.07.007. Epub 2022 Aug 16. Trends Cell Biol. 2023. PMID: 35981909 Review.
Cited by
-
Clinical Relevance of Secreted Small Noncoding RNAs in an Embryo Implantation Potential Prediction at Morula and Blastocyst Development Stages.Life (Basel). 2021 Dec 1;11(12):1328. doi: 10.3390/life11121328. Life (Basel). 2021. PMID: 34947859 Free PMC article.
-
Cell-free Translation: Preparation and Validation of Translation-competent Extracts from Saccharomyces cerevisiae.Bio Protoc. 2021 Sep 20;11(18):e4093. doi: 10.21769/BioProtoc.4093. eCollection 2021 Sep 20. Bio Protoc. 2021. PMID: 34692902 Free PMC article.
-
Polymer Physics Models Reveal Structural Folding Features of Single-Molecule Gene Chromatin Conformations.Int J Mol Sci. 2024 Sep 23;25(18):10215. doi: 10.3390/ijms251810215. Int J Mol Sci. 2024. PMID: 39337699 Free PMC article.
-
The ribosome-associated chaperone Zuo1 controls translation upon TORC1 inhibition.EMBO J. 2023 Dec 11;42(24):e113240. doi: 10.15252/embj.2022113240. Epub 2023 Nov 20. EMBO J. 2023. PMID: 37984430 Free PMC article.
-
Specific branches of the proteostasis network regulate the toxicity associated with mistranslation.Nucleic Acids Res. 2025 May 10;53(9):gkaf428. doi: 10.1093/nar/gkaf428. Nucleic Acids Res. 2025. PMID: 40377218 Free PMC article.
References
-
- Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C.C., Li G.-W., Zhou S., King D., Shen P.S., Weibezahn J., et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

