Cofactor molecules: Essential partners for infectious prions
- PMID: 32958241
- PMCID: PMC7768309
- DOI: 10.1016/bs.pmbts.2020.07.009
Cofactor molecules: Essential partners for infectious prions
Abstract
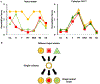
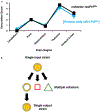
The protein-only hypothesis predicts that infectious mammalian prions are composed solely of PrPSc, a misfolded conformer of the normal prion protein, PrPC. However, to date, all wild type protein-only PrPSc preparations lack significant levels of prion infectivity. Using a systemic biochemical approach, our laboratory isolated and identified two different endogenous cofactor molecules, RNA (Deleault et al., 2003 [50]; Deleault et al., 2007 [59]) and phosphatidylethanolamine (Deleault et al., 2012 [61]; Deleault et al., 2012 [18]), which facilitate the formation of prions with high levels of specific infectivity, leading us to propose to the alternative hypothesis that cofactor molecules are required to form wild type infectious prions (Deleault et al., 2007 [59]; Deleault et al., 2012 [18]; Geoghegan et al., 2007 [57]). In addition, we found that purified cofactor molecules restrict the strain properties of chemically defined infectious prions (Deleault et al., 2012 [18]), suggesting a "cofactor selection" model in which natural variation in the distribution of strain-specific cofactor molecules in different parts of the brain may be responsible for strain-dependent patterns of neurotropism (Deleault et al., 2012 [18]; Geoghegan et al., 2007 [57]).
Keywords: Cofactor; Cofactor selection; Mammalian; Phosphatidylethanolamine; Phospholipid; Polyanion; Prion; RNA; Specific infectivity.
© 2020 Elsevier Inc. All rights reserved.
Figures
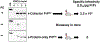
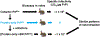
Similar articles
-
Full restoration of specific infectivity and strain properties from pure mammalian prion protein.PLoS Pathog. 2019 Mar 25;15(3):e1007662. doi: 10.1371/journal.ppat.1007662. eCollection 2019 Mar. PLoS Pathog. 2019. PMID: 30908557 Free PMC article.
-
Synthesis of high titer infectious prions with cofactor molecules.J Biol Chem. 2014 Jul 18;289(29):19850-4. doi: 10.1074/jbc.R113.511329. Epub 2014 May 23. J Biol Chem. 2014. PMID: 24860097 Free PMC article. Review.
-
Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions.Proc Natl Acad Sci U S A. 2012 Jul 10;109(28):E1938-46. doi: 10.1073/pnas.1206999109. Epub 2012 Jun 18. Proc Natl Acad Sci U S A. 2012. PMID: 22711839 Free PMC article.
-
Elucidating the role of cofactors in mammalian prion propagation.Prion. 2014 Jan-Feb;8(1):100-5. doi: 10.4161/pri.27501. Prion. 2014. PMID: 24365977 Free PMC article. Review.
-
Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids.Proc Natl Acad Sci U S A. 2012 May 29;109(22):8546-51. doi: 10.1073/pnas.1204498109. Epub 2012 May 14. Proc Natl Acad Sci U S A. 2012. PMID: 22586108 Free PMC article.
Cited by
-
Cryo-EM of prion strains from the same genotype of host identifies conformational determinants.PLoS Pathog. 2022 Nov 7;18(11):e1010947. doi: 10.1371/journal.ppat.1010947. eCollection 2022 Nov. PLoS Pathog. 2022. PMID: 36342968 Free PMC article.
-
RNA-seq analyses reveal the relevance of RNAs involved in ribosomal complex to induce mammalian prion protein aggregation and phase separation in vitro.RNA Biol. 2025 Dec;22(1):1-16. doi: 10.1080/15476286.2025.2508107. Epub 2025 May 29. RNA Biol. 2025. PMID: 40438940 Free PMC article.
-
Propagation and Dissemination Strategies of Transmissible Spongiform Encephalopathy Agents in Mammalian Cells.Int J Mol Sci. 2022 Mar 8;23(6):2909. doi: 10.3390/ijms23062909. Int J Mol Sci. 2022. PMID: 35328330 Free PMC article. Review.
-
Prion strains: shining new light on old concepts.Cell Tissue Res. 2023 Apr;392(1):113-133. doi: 10.1007/s00441-022-03665-2. Epub 2022 Jul 7. Cell Tissue Res. 2023. PMID: 35796874 Free PMC article. Review.
-
Conformational diversity in purified prions produced in vitro.PLoS Pathog. 2023 Jan 10;19(1):e1011083. doi: 10.1371/journal.ppat.1011083. eCollection 2023 Jan. PLoS Pathog. 2023. PMID: 36626391 Free PMC article.
References
-
- Bessen RA, Marsh RF. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J Gen Virol. 1992;73(pt 2):329–334. - PubMed
-
- Loftus B, Rogers M. Characterization of a prion protein (PrP) gene from rabbit; a species with apparent resistance to infection by prions. Gene. 1997;184(2):215–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

