Convergent molecular, cellular, and cortical neuroimaging signatures of major depressive disorder
- PMID: 32958675
- PMCID: PMC7547155
- DOI: 10.1073/pnas.2008004117
Convergent molecular, cellular, and cortical neuroimaging signatures of major depressive disorder
Abstract
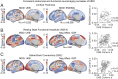
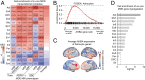
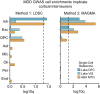
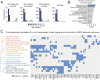
Major depressive disorder emerges from the complex interactions of biological systems that span genes and molecules through cells, networks, and behavior. Establishing how neurobiological processes coalesce to contribute to depression requires a multiscale approach, encompassing measures of brain structure and function as well as genetic and cell-specific transcriptional data. Here, we examine anatomical (cortical thickness) and functional (functional variability, global brain connectivity) correlates of depression and negative affect across three population-imaging datasets: UK Biobank, Brain Genomics Superstruct Project, and Enhancing NeuroImaging through Meta Analysis (ENIGMA; combined n ≥ 23,723). Integrative analyses incorporate measures of cortical gene expression, postmortem patient transcriptional data, depression genome-wide association study (GWAS), and single-cell gene transcription. Neuroimaging correlates of depression and negative affect were consistent across three independent datasets. Linking ex vivo gene down-regulation with in vivo neuroimaging, we find that transcriptional correlates of depression imaging phenotypes track gene down-regulation in postmortem cortical samples of patients with depression. Integrated analysis of single-cell and Allen Human Brain Atlas expression data reveal somatostatin interneurons and astrocytes to be consistent cell associates of depression, through both in vivo imaging and ex vivo cortical gene dysregulation. Providing converging evidence for these observations, GWAS-derived polygenic risk for depression was enriched for genes expressed in interneurons, but not glia. Underscoring the translational potential of multiscale approaches, the transcriptional correlates of depression-linked brain function and structure were enriched for disorder-relevant molecular pathways. These findings bridge levels to connect specific genes, cell classes, and biological pathways to in vivo imaging correlates of depression.
Keywords: astrocytes; gene expression; major depressive disorder; neuroimaging; somatostatin interneurons.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
Competing interest statement: A.M.C. holds equity in Spring Care Inc, Fitbit Inc, and UnitedHealthcare Inc. He is lead inventor on three patent submissions relating to treatment for major depressive disorder (US Patent and Trademark Office [USPTO] docket no. Y0087.70116US00, USPTO provisional application no. 62/491,660, and USPTO provisional application no. 62/629,041). He has consulted for Fortress Biotech on antidepressant drug development.
Figures


References
-
- Sullivan P. F., Neale M. C., Kendler K. S., Genetic epidemiology of major depression: Review and meta-analysis. Am. J. Psychiatry 157, 1552–1562 (2000). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

