Rational programming of history-dependent logic in cellular populations
- PMID: 32958811
- PMCID: PMC7506022
- DOI: 10.1038/s41467-020-18455-z
Rational programming of history-dependent logic in cellular populations
Abstract
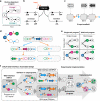
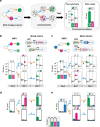
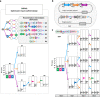
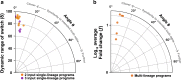
Genetic programs operating in a history-dependent fashion are ubiquitous in nature and govern sophisticated processes such as development and differentiation. The ability to systematically and predictably encode such programs would advance the engineering of synthetic organisms and ecosystems with rich signal processing abilities. Here we implement robust, scalable history-dependent programs by distributing the computational labor across a cellular population. Our design is based on standardized recombinase-driven DNA scaffolds expressing different genes according to the order of occurrence of inputs. These multicellular computing systems are highly modular, do not require cell-cell communication channels, and any program can be built by differential composition of strains containing well-characterized logic scaffolds. We developed automated workflows that researchers can use to streamline program design and optimization. We anticipate that the history-dependent programs presented here will support many applications using cellular populations for material engineering, biomanufacturing and healthcare.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
An Automated Design Framework for Multicellular Recombinase Logic.ACS Synth Biol. 2018 May 18;7(5):1406-1412. doi: 10.1021/acssynbio.8b00016. Epub 2018 May 4. ACS Synth Biol. 2018. PMID: 29641183 Free PMC article.
-
Hierarchical composition of reliable recombinase logic devices.Nat Commun. 2019 Jan 28;10(1):456. doi: 10.1038/s41467-019-08391-y. Nat Commun. 2019. PMID: 30692530 Free PMC article.
-
Computational Methods for the Design of Recombinase Logic Circuits with Adaptable Circuit Specifications.Methods Mol Biol. 2023;2553:155-171. doi: 10.1007/978-1-0716-2617-7_8. Methods Mol Biol. 2023. PMID: 36227543
-
Engineering multicellular traits in synthetic microbial populations.Curr Opin Chem Biol. 2012 Aug;16(3-4):370-8. doi: 10.1016/j.cbpa.2012.04.002. Epub 2012 May 15. Curr Opin Chem Biol. 2012. PMID: 22591687 Review.
-
Synthesizing biomolecule-based Boolean logic gates.ACS Synth Biol. 2013 Feb 15;2(2):72-82. doi: 10.1021/sb3001112. ACS Synth Biol. 2013. PMID: 23526588 Free PMC article. Review.
Cited by
-
A rapid and standardized workflow for functional assessment of bacterial biosensors in fecal samples.Front Bioeng Biotechnol. 2022 Aug 22;10:859600. doi: 10.3389/fbioe.2022.859600. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36072290 Free PMC article.
-
Decoding and recoding plant development.Plant Physiol. 2021 Oct 5;187(2):515-526. doi: 10.1093/plphys/kiab336. Plant Physiol. 2021. PMID: 35237818 Free PMC article. Review.
-
Synthetic biology in the clinic: engineering vaccines, diagnostics, and therapeutics.Cell. 2021 Feb 18;184(4):881-898. doi: 10.1016/j.cell.2021.01.017. Epub 2021 Feb 10. Cell. 2021. PMID: 33571426 Free PMC article. Review.
-
Next generation synthetic memory via intercepting recombinase function.Nat Commun. 2023 Aug 29;14(1):5255. doi: 10.1038/s41467-023-41043-w. Nat Commun. 2023. PMID: 37644045 Free PMC article.
-
An integrase toolbox to record gene-expression during plant development.Nat Commun. 2023 Apr 3;14(1):1844. doi: 10.1038/s41467-023-37607-5. Nat Commun. 2023. PMID: 37012288 Free PMC article.
References
-
- Anderson PW. More is different. Science. 1972;177:393–396. - PubMed
-
- Doebeli M, Ispolatov I. Complexity and diversity. Science. 2010;328:494–497. - PubMed
-
- Durand PM, Sym S, Michod RE. Programmed cell death and complexity in microbial systems. Curr. Biol. 2016;26:R587–R593. - PubMed
-
- Wolpert L. Positional information and pattern formation. Curr. Top. Dev. Biol. 2016;117:597–608. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

