Lung mesenchymal cells elicit lipid storage in neutrophils that fuel breast cancer lung metastasis
- PMID: 32958928
- PMCID: PMC7584447
- DOI: 10.1038/s41590-020-0783-5
Lung mesenchymal cells elicit lipid storage in neutrophils that fuel breast cancer lung metastasis
Abstract
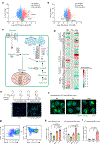
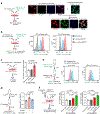
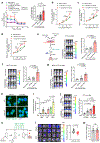
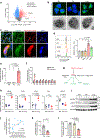
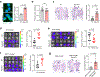
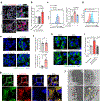
Acquisition of a lipid-laden phenotype by immune cells has been defined in infectious diseases and atherosclerosis but remains largely uncharacterized in cancer. Here, in breast cancer models, we found that neutrophils are induced to accumulate neutral lipids upon interaction with resident mesenchymal cells in the premetastatic lung. Lung mesenchymal cells elicit this process through repressing the adipose triglyceride lipase (ATGL) activity in neutrophils in prostaglandin E2-dependent and -independent manners. In vivo, neutrophil-specific deletion of genes encoding ATGL or ATGL inhibitory factors altered neutrophil lipid profiles and breast tumor lung metastasis in mice. Mechanistically, lipids stored in lung neutrophils are transported to metastatic tumor cells through a macropinocytosis-lysosome pathway, endowing tumor cells with augmented survival and proliferative capacities. Pharmacological inhibition of macropinocytosis significantly reduced metastatic colonization by breast tumor cells in vivo. Collectively, our work reveals that neutrophils serve as an energy reservoir to fuel breast cancer lung metastasis.
Figures

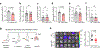





References
-
- Liu Y, Cao X. Characteristics and Significance of the Pre-metastatic Niche. Cancer cell 2016, 30(5): 668–681. - PubMed
-
- Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, et al. Pre-metastatic niches: organ-specific homes for metastases. Nature reviews Cancer 2017, 17(5): 302–317. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA034196/CA/NCI NIH HHS/United States
- R37 CA237307/CA/NCI NIH HHS/United States
- R35 GM133562/GM/NIGMS NIH HHS/United States
- R01-AI132963/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)/International
- R00 CA188093/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

