Identification of common differentially expressed genes in Turner (45,X) and Klinefelter (47,XXY) syndromes using bioinformatics analysis
- PMID: 32959501
- PMCID: PMC7667333
- DOI: 10.1002/mgg3.1503
Identification of common differentially expressed genes in Turner (45,X) and Klinefelter (47,XXY) syndromes using bioinformatics analysis
Abstract
Background: Analysis of patients with chromosomal abnormalities, including Turner syndrome and Klinefelter syndrome, has highlighted the importance of X-linked gene dosage as a contributing factor for disease susceptibility. Escape from X-inactivation and X-linked imprinting can result in transcriptional differences between normal men and women as well as in patients with sex chromosome abnormalities.
Objective: To identify differentially expressed genes among patients with Turner (45,X) and Klinefelter (46,XXY) syndrome using bioinformatics analysis.
Methodology: Two gene expression data sets of Turner (45,X) and Klinefelter syndrome (47,XXY) were obtained from the Gene Omnibus Expression (GEO) database of the National Center for Biotechnology Information (NCBI). Statistical analysis was performed using R Bioconductor libraries. Differentially expressed genes (DEGs) were determined using significance analysis of microarray (SAM). The functional annotation of the DEGs was performed with DAVID v6.8 (The Database for Annotation, Visualization, and Integrated Discovery).
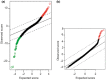
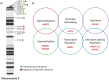
Results: There are no genes over-expressed simultaneously in both diseases. However, when crossing the list of under-expressed genes for 45,X cells and the list of over-expressed genes for 47,XXY cells, there are 16 common genes: SLC25A6, AKAP17A, ASMTL, KDM5C, KDM6A, ATRX, CSF2RA, DHRSX, CD99, ZBED1, EIF1AX, MVB12B, SMC1A, P2RY8, DOCK7, DDX3X, eight of which are involved in the regulation of gene expression by epigenetic mechanisms, regulation of splicing processes and protein synthesis.
Conclusion: Of the 16 identified as under-expressed in 45,X cells and over-expressed in 47,XXY cells, 14 are located in X chromosome and 2 in autosomal chromosome; 8 of these genes are involved in the regulation of gene expression: 5 genes are related to epigenetic mechanisms, 2 in regulation of splicing processes, and 1 in the protein synthesis process. Our results are limited by it being the product of a bioinformatic analysis from mRNA isolated from whole blood, this makes necessary further exploration of the relationships between these genes and Turner syndrome and Klinefelter syndrome in the future.
© 2020 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals LLC.
Conflict of interest statement
None declared.
Figures

Similar articles
-
Integrated functional genomic analyses of Klinefelter and Turner syndromes reveal global network effects of altered X chromosome dosage.Proc Natl Acad Sci U S A. 2020 Mar 3;117(9):4864-4873. doi: 10.1073/pnas.1910003117. Epub 2020 Feb 18. Proc Natl Acad Sci U S A. 2020. PMID: 32071206 Free PMC article.
-
Epigenetic and transcriptomic consequences of excess X-chromosome material in 47,XXX syndrome-A comparison with Turner syndrome and 46,XX females.Am J Med Genet C Semin Med Genet. 2020 Jun;184(2):279-293. doi: 10.1002/ajmg.c.31799. Epub 2020 Jun 3. Am J Med Genet C Semin Med Genet. 2020. PMID: 32489015
-
Bioinformatic Analysis Identifies Potential Key Genes in the Pathogenesis of Turner Syndrome.Front Endocrinol (Lausanne). 2020 Mar 6;11:104. doi: 10.3389/fendo.2020.00104. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 32210915 Free PMC article.
-
The contribution of Xp22.31 gene dosage to Turner and Klinefelter syndromes and sex-biased phenotypes.Eur J Med Genet. 2021 Apr;64(4):104169. doi: 10.1016/j.ejmg.2021.104169. Epub 2021 Feb 19. Eur J Med Genet. 2021. PMID: 33610733 Review.
-
Chromosomal Aberrations with Endocrine Relevance (Turner Syndrome, Klinefelter Syndrome, Prader-Willi Syndrome).Exp Suppl. 2019;111:443-473. doi: 10.1007/978-3-030-25905-1_20. Exp Suppl. 2019. PMID: 31588543 Review.
Cited by
-
Dosage of the pseudoautosomal gene SLC25A6 is implicated in QTc interval duration.Sci Rep. 2023 Jul 26;13(1):12089. doi: 10.1038/s41598-023-38867-3. Sci Rep. 2023. PMID: 37495650 Free PMC article.
-
A Review of Recent Developments in Turner Syndrome Research.J Cardiovasc Dev Dis. 2021 Oct 23;8(11):138. doi: 10.3390/jcdd8110138. J Cardiovasc Dev Dis. 2021. PMID: 34821691 Free PMC article. Review.
-
Procoagulant Imbalance in Klinefelter Syndrome Assessed by Thrombin Generation Assay and Whole-Blood Thromboelastometry.J Clin Endocrinol Metab. 2021 Mar 25;106(4):e1660-e1672. doi: 10.1210/clinem/dgaa936. J Clin Endocrinol Metab. 2021. PMID: 33382882 Free PMC article.
-
The transcriptomic landscape of monosomy X (45,X) during early human fetal and placental development.Commun Biol. 2025 Feb 16;8(1):249. doi: 10.1038/s42003-025-07699-4. Commun Biol. 2025. PMID: 39956831 Free PMC article.
References
-
- Alberts, B. (2007). Molecular biology of the cell (5th ed.). New York: 10.1249/mss.0b013e318185ce9d - DOI
-
- Allis, D.C. , Caparros, M.-L. , Jenuwein, T. , & Reinberg, D. (2015). Epigenetics (2nd ed.). New York: Retrieved from https://cshlpress.com/default.tpl?cart=15832879119832666&action=fu...
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

