Coupling 193 nm Ultraviolet Photodissociation and Ion Mobility for Sequence Characterization of Conformationally-Selected Peptides
- PMID: 32959654
- PMCID: PMC8127984
- DOI: 10.1021/jasms.0c00259
Coupling 193 nm Ultraviolet Photodissociation and Ion Mobility for Sequence Characterization of Conformationally-Selected Peptides
Abstract
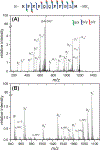
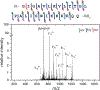
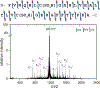
Ultraviolet photodissociation (UVPD) has emerged as a useful technique for characterizing peptide, protein, and protein complex primary and secondary structure. 193 nm UVPD, specifically, enables extensive covalent fragmentation of the peptide backbone without the requirement of a specific side chain chromophore and with no precursor charge state dependence. We have modified a commercial quadrupole-ion mobility-time-of-flight (Q-IM-TOF) mass spectrometer to include 193 nm UVPD following ion mobility. Ion mobility (IM) is a gas-phase separation technique that enables separation of ions by their size, shape, and charge, providing an orthogonal dimension of separation to mass analysis. Following instrument modifications, we characterized the performance of, and information that could be generated from, this new setup using the model peptides substance P, melittin, and insulin chain B. These experiments show extensive fragmentation across the peptide backbone and a variety of ion types as expected from 193 nm UVPD. Additionally, y-2 ions (along with complementary a+2 and b+2 ions) N-terminal to proline were observed. Combining the IM separation and mobility gating capabilities with UVPD, we demonstrate the ability to accomplish both mass- and mobility-selection of bradykinin des-Arg9 and des-Arg1 peptides followed by complete sequence characterization by UVPD. The new capabilities of this modified instrument demonstrate the utility of combining IM with UVPD because isobaric species cannot be independently selected with a traditional quadrupole alone.
Conflict of interest statement
The authors declare the following financial competing interest(s): Jeffery Brown is an employee of Waters Corporation, which manufactures and sells Synapt G2-S instruments.
Figures

Similar articles
-
Use of Ultraviolet Photodissociation Coupled with Ion Mobility Mass Spectrometry To Determine Structure and Sequence from Drift Time Selected Peptides and Proteins.Anal Chem. 2016 Oct 18;88(20):9964-9971. doi: 10.1021/acs.analchem.6b01705. Epub 2016 Oct 3. Anal Chem. 2016. PMID: 27631466
-
[Vacuum ultraviolet laser dissociation and proteomic analysis of halogenated peptides].Se Pu. 2025 Feb;43(2):131-138. doi: 10.3724/SP.J.1123.2024.08009. Se Pu. 2025. PMID: 39844703 Free PMC article. Chinese.
-
MS/MS simplification by 355 nm ultraviolet photodissociation of chromophore-derivatized peptides in a quadrupole ion trap.Anal Chem. 2007 Oct 15;79(20):7883-92. doi: 10.1021/ac071241t. Epub 2007 Sep 11. Anal Chem. 2007. PMID: 17845006
-
Ultraviolet Photodissociation Mass Spectrometry for Analysis of Biological Molecules.Chem Rev. 2020 Apr 8;120(7):3328-3380. doi: 10.1021/acs.chemrev.9b00440. Epub 2019 Dec 18. Chem Rev. 2020. PMID: 31851501 Free PMC article. Review.
-
Photodissociation mass spectrometry: new tools for characterization of biological molecules.Chem Soc Rev. 2014 Apr 21;43(8):2757-83. doi: 10.1039/c3cs60444f. Epub 2014 Jan 30. Chem Soc Rev. 2014. PMID: 24481009 Free PMC article. Review.
Cited by
-
Characterization of the T4 gp32-ssDNA complex by native, cross-linking, and ultraviolet photodissociation mass spectrometry.Chem Sci. 2021 Sep 23;12(41):13764-13776. doi: 10.1039/d1sc02861h. eCollection 2021 Oct 27. Chem Sci. 2021. PMID: 34760161 Free PMC article.
-
Instrumentation at the Leading Edge of Proteomics.Anal Chem. 2024 May 21;96(20):7976-8010. doi: 10.1021/acs.analchem.3c04497. Epub 2024 May 13. Anal Chem. 2024. PMID: 38738990 Free PMC article. Review.
-
Surface-induced Dissociation Mass Spectrometry as a Structural Biology Tool.Chem Rev. 2022 Apr 27;122(8):7442-7487. doi: 10.1021/acs.chemrev.1c00309. Epub 2021 Nov 2. Chem Rev. 2022. PMID: 34726898 Free PMC article. Review.
-
Trapped Ion Mobility Spectrometry, Ultraviolet Photodissociation, and Time-of-Flight Mass Spectrometry for Gas-Phase Peptide Isobars/Isomers/Conformers Discrimination.J Am Soc Mass Spectrom. 2022 Jul 6;33(7):1267-1275. doi: 10.1021/jasms.2c00091. Epub 2022 Jun 5. J Am Soc Mass Spectrom. 2022. PMID: 35658468 Free PMC article.
-
Tandem-trapped ion mobility spectrometry/mass spectrometry (tTIMS/MS): a promising analytical method for investigating heterogenous samples.Analyst. 2022 May 30;147(11):2317-2337. doi: 10.1039/d2an00335j. Analyst. 2022. PMID: 35521797 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

