Influenza A virus co-opts ERI1 exonuclease bound to histone mRNA to promote viral transcription
- PMID: 32960265
- PMCID: PMC7544206
- DOI: 10.1093/nar/gkaa771
Influenza A virus co-opts ERI1 exonuclease bound to histone mRNA to promote viral transcription
Abstract
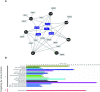
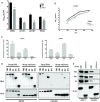
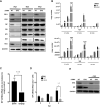
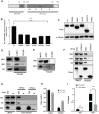
Cellular exonucleases involved in the processes that regulate RNA stability and quality control have been shown to restrict or to promote the multiplication cycle of numerous RNA viruses. Influenza A viruses are major human pathogens that are responsible for seasonal epidemics, but the interplay between viral proteins and cellular exonucleases has never been specifically studied. Here, using a stringent interactomics screening strategy and an siRNA-silencing approach, we identified eight cellular factors among a set of 75 cellular proteins carrying exo(ribo)nuclease activities or involved in RNA decay processes that support influenza A virus multiplication. We show that the exoribonuclease ERI1 interacts with the PB2, PB1 and NP components of the viral ribonucleoproteins and is required for viral mRNA transcription. More specifically, we demonstrate that the protein-protein interaction is RNA dependent and that both the RNA binding and exonuclease activities of ERI1 are required to promote influenza A virus transcription. Finally, we provide evidence that during infection, the SLBP protein and histone mRNAs co-purify with vRNPs alongside ERI1, indicating that ERI1 is most probably recruited when it is present in the histone pre-mRNA processing complex in the nucleus.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
The Cellular Factor NXP2/MORC3 Is a Positive Regulator of Influenza Virus Multiplication.J Virol. 2015 Oct;89(19):10023-30. doi: 10.1128/JVI.01530-15. Epub 2015 Jul 22. J Virol. 2015. PMID: 26202233 Free PMC article.
-
A Nucleolar Protein, Ribosomal RNA Processing 1 Homolog B (RRP1B), Enhances the Recruitment of Cellular mRNA in Influenza Virus Transcription.J Virol. 2015 Nov;89(22):11245-55. doi: 10.1128/JVI.01487-15. Epub 2015 Aug 26. J Virol. 2015. PMID: 26311876 Free PMC article.
-
The Nucleolar Protein LYAR Facilitates Ribonucleoprotein Assembly of Influenza A Virus.J Virol. 2018 Nov 12;92(23):e01042-18. doi: 10.1128/JVI.01042-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30209172 Free PMC article.
-
Biogenesis, assembly, and export of viral messenger ribonucleoproteins in the influenza A virus infected cell.RNA Biol. 2013 Aug;10(8):1274-82. doi: 10.4161/rna.25356. Epub 2013 Jun 17. RNA Biol. 2013. PMID: 23807439 Free PMC article. Review.
-
[Molecular mechanism of replication and transcription of the influenza virus genome and host factors].Uirusu. 2006 Jun;56(1):99-108. doi: 10.2222/jsv.56.99. Uirusu. 2006. PMID: 17038818 Review. Japanese.
Cited by
-
Functional interactomes of the Ebola virus polymerase identified by proximity proteomics in the context of viral replication.Cell Rep. 2022 Mar 22;38(12):110544. doi: 10.1016/j.celrep.2022.110544. Cell Rep. 2022. PMID: 35320713 Free PMC article.
-
hnRNPM regulates influenza A virus replication through distinct mechanisms in human and avian cells: implications for cross-species transmission.J Virol. 2025 Jun 17;99(6):e0006725. doi: 10.1128/jvi.00067-25. Epub 2025 May 28. J Virol. 2025. PMID: 40434105 Free PMC article.
-
Mendelian randomization provides a multi-omics perspective on the regulation of genes involved in ribosome biogenesis in relation to cardiac structure and function.Clin Epigenetics. 2025 Mar 5;17(1):42. doi: 10.1186/s13148-025-01850-y. Clin Epigenetics. 2025. PMID: 40045424 Free PMC article.
-
Impact of Ribosome Activity on SARS-CoV-2 LNP - Based mRNA Vaccines.Front Mol Biosci. 2021 Apr 20;8:654866. doi: 10.3389/fmolb.2021.654866. eCollection 2021. Front Mol Biosci. 2021. PMID: 33959636 Free PMC article. Review.
-
Identification of a 6-RBP gene signature for a comprehensive analysis of glioma and ischemic stroke: Cognitive impairment and aging-related hypoxic stress.Front Aging Neurosci. 2022 Sep 1;14:951197. doi: 10.3389/fnagi.2022.951197. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36118697 Free PMC article.
References
-
- Łabno A., Tomecki R., Dziembowski A.. Cytoplasmic RNA decay pathways - enzymes and mechanisms. Biochim. Biophys. Acta - Mol. Cell Res. 2016; 1863:3125–3147. - PubMed
-
- Mugridge J.S., Coller J., Gross J.D.. Structural and molecular mechanisms for the control of eukaryotic 5′–3′ mRNA decay. Nat. Struct. Mol. Biol. 2018; 25:1077–1085. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

