Collision-Induced Unfolding Differentiates Functional Variants of the KCNQ1 Voltage Sensor Domain
- PMID: 32960579
- PMCID: PMC8106873
- DOI: 10.1021/jasms.0c00288
Collision-Induced Unfolding Differentiates Functional Variants of the KCNQ1 Voltage Sensor Domain
Abstract
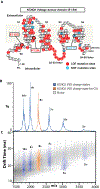
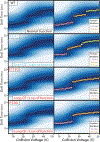
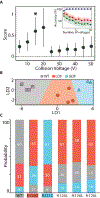
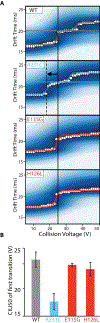
The KCNQ1 voltage-gated potassium channel regulates the repolarization of cardiac cells, and a plurality of point mutations in its voltage-sensing domain (VSD) are associated with toxic gain or loss of pore function, resulting in disease. As is the case with many disease-associated membrane proteins, there are hundreds of human variants of interest identified for KCNQ1; however, a significant portion of these variants have not been characterized in relation to their functional and disease associations. Additionally, as the VSD consists of four transmembrane helices, studies into dynamic structural differences among KCNQ1 VSD variants are hindered by the current limitations and deficits in the high-resolution structure determination of membrane proteins. Here, we use native ion mobility-mass spectrometry and collision-induced unfolding (CIU) to address the need for a high throughput-compatible method for the structural characterization of membrane protein variants of unknown significance using the KCNQ1 VSD as a model system. We perform CIU on wild-type and three mutant KCNQ1 VSD forms associated with the toxic gain or loss of function and show through both automated feature detection and comprehensive difference analysis of the CIU data sets that the variants are clearly grouped by function and disease association. We also construct a classification scheme based on the CIU data sets, which is able to differentiate the variant functional groups and classify a recently characterized variant to its correct grouping. Further, we probe the stability of the KCNQ1 VSD variants when liberated from C12E8 micelles at pH 8.0 and find preliminary evidence that the R231C mutation associated with the gain of the pore function is destabilized relative to the wild-type and loss of function variants.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Escribá PV; González-Ros JM; Goñi FM; Kinnunen PKJ; Vigh L; Sánchez-Magraner L; Fernández AM; Busquets X; Horváth I; Barceló-Coblijn G Membranes: A Meeting Point for Lipids, Proteins and Therapies: Translational Medicine. J. Cell. Mol. Med 2008, 12 (3), 829–875. 10.1111/j.1582-4934.2008.00281.x. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

