Conserved actin machinery drives microtubule-independent motility and phagocytosis in Naegleria
- PMID: 32960946
- PMCID: PMC7594500
- DOI: 10.1083/jcb.202007158
Conserved actin machinery drives microtubule-independent motility and phagocytosis in Naegleria
Abstract
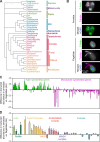
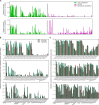
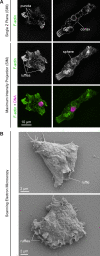
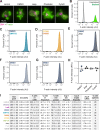
Much of our understanding of actin-driven phenotypes in eukaryotes has come from the "yeast-to-human" opisthokont lineage and the related amoebozoa. Outside of these groups lies the genus Naegleria, which shared a common ancestor with humans >1 billion years ago and includes the "brain-eating amoeba." Unlike nearly all other known eukaryotic cells, Naegleria amoebae lack interphase microtubules; this suggests that actin alone drives phenotypes like cell crawling and phagocytosis. Naegleria therefore represents a powerful system to probe actin-driven functions in the absence of microtubules, yet surprisingly little is known about its actin cytoskeleton. Using genomic analysis, microscopy, and molecular perturbations, we show that Naegleria encodes conserved actin nucleators and builds Arp2/3-dependent lamellar protrusions. These protrusions correlate with the capacity to migrate and eat bacteria. Because human cells also use Arp2/3-dependent lamellar protrusions for motility and phagocytosis, this work supports an evolutionarily ancient origin for these processes and establishes Naegleria as a natural model system for studying microtubule-independent cytoskeletal phenotypes.
© 2020 Velle and Fritz-Laylin.
Figures










Comment in
-
Moving simply: Naegleria crawls and feeds using an ancient Arp2/3-dependent mechanism.J Cell Biol. 2020 Nov 2;219(11):e202009031. doi: 10.1083/jcb.202009031. J Cell Biol. 2020. PMID: 33064835 Free PMC article.
Similar articles
-
Moving simply: Naegleria crawls and feeds using an ancient Arp2/3-dependent mechanism.J Cell Biol. 2020 Nov 2;219(11):e202009031. doi: 10.1083/jcb.202009031. J Cell Biol. 2020. PMID: 33064835 Free PMC article.
-
Morphodynamics of the Actin-Rich Cytoskeleton in Entamoeba histolytica.Front Cell Infect Microbiol. 2018 May 29;8:179. doi: 10.3389/fcimb.2018.00179. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29896453 Free PMC article.
-
From Molecules to Amoeboid Movement: A New Way for Understanding the Morphology Through Actin-Binding Proteins.Biomolecules. 2024 Dec 11;14(12):1583. doi: 10.3390/biom14121583. Biomolecules. 2024. PMID: 39766290 Free PMC article.
-
New twists in actin-microtubule interactions.Mol Biol Cell. 2021 Feb 1;32(3):211-217. doi: 10.1091/mbc.E19-09-0491. Mol Biol Cell. 2021. PMID: 33507109 Free PMC article. Review.
-
Bacterial subversion of host cytoskeletal machinery: hijacking formins and the Arp2/3 complex.Bioessays. 2014 Jul;36(7):687-96. doi: 10.1002/bies.201400038. Epub 2014 May 21. Bioessays. 2014. PMID: 24849003 Review.
Cited by
-
An Atypical F-Actin Capping Protein Modulates Cytoskeleton Behaviors Crucial for Trichomonas vaginalis Colonization.Microbiol Spectr. 2023 Aug 17;11(4):e0059623. doi: 10.1128/spectrum.00596-23. Epub 2023 Jun 13. Microbiol Spectr. 2023. PMID: 37310229 Free PMC article.
-
Computational discovery of potential therapeutic agents against brain-eating amoeba (Naegleria fowleri).PLoS One. 2025 Jul 11;20(7):e0327621. doi: 10.1371/journal.pone.0327621. eCollection 2025. PLoS One. 2025. PMID: 40644400 Free PMC article.
-
Endocytosis in cancer and cancer therapy.Nat Rev Cancer. 2023 Jul;23(7):450-473. doi: 10.1038/s41568-023-00574-6. Epub 2023 May 22. Nat Rev Cancer. 2023. PMID: 37217781 Review.
-
Actin network evolution as a key driver of eukaryotic diversification.J Cell Sci. 2024 Aug 1;137(15):jcs261660. doi: 10.1242/jcs.261660. Epub 2024 Aug 9. J Cell Sci. 2024. PMID: 39120594 Free PMC article. Review.
-
Naegleria genus pangenome reveals new structural and functional insights into the versatility of these free-living amoebae.Front Microbiol. 2023 Feb 1;13:1056418. doi: 10.3389/fmicb.2022.1056418. eCollection 2022. Front Microbiol. 2023. PMID: 36817109 Free PMC article.
References
-
- Abercrombie M. 1980. The Croonian Lecture, 1978- The crawling movement of metazoan cells. Proc. R. Soc. Lond. B Biol. Sci. 207:129–147. 10.1098/rspb.1980.0017 - DOI
-
- Baggett A.W., Cournia Z., Han M.S., Patargias G., Glass A.C., Liu S.Y., and Nolen B.J.. 2012. Structural characterization and computer-aided optimization of a small-molecule inhibitor of the Arp2/3 complex, a key regulator of the actin cytoskeleton. ChemMedChem. 7:1286–1294. 10.1002/cmdc.201200104 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

