Deconstructing Stepwise Fate Conversion of Human Fibroblasts to Neurons by MicroRNAs
- PMID: 32961143
- PMCID: PMC7796891
- DOI: 10.1016/j.stem.2020.08.015
Deconstructing Stepwise Fate Conversion of Human Fibroblasts to Neurons by MicroRNAs
Abstract
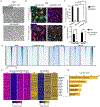
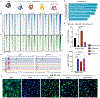
Cell-fate conversion generally requires reprogramming effectors to both introduce fate programs of the target cell type and erase the identity of starting cell population. Here, we reveal insights into the activity of microRNAs miR-9/9∗ and miR-124 (miR-9/9∗-124) as reprogramming agents that orchestrate direct conversion of human fibroblasts into motor neurons by first eradicating fibroblast identity and promoting uniform transition to a neuronal state in sequence. We identify KLF-family transcription factors as direct target genes for miR-9/9∗-124 and show their repression is critical for erasing fibroblast fate. Subsequent gain of neuronal identity requires upregulation of a small nuclear RNA, RN7SK, which induces accessibilities of chromatin regions and neuronal gene activation to push cells to a neuronal state. Our study defines deterministic components in the microRNA-mediated reprogramming cascade.
Keywords: cell fate; chromatin regulation; direct conversion; epigenetics; microRNA; neuronal reprogramming; non-coding RNA; single-cell RNA-sequencing.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures



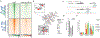

Comment in
-
One Big Step to a Neuron, Two Small Steps for miRNAs.Cell Stem Cell. 2021 Jan 7;28(1):1-2. doi: 10.1016/j.stem.2020.12.007. Cell Stem Cell. 2021. PMID: 33417865
References
-
- Bazi Z, Bertacchi M, Abasi M, Mohammadi-Yeganeh S, Soleimani M, Wagner N, and Ghanbarian H (2018). Rn7SK small nuclear RNA is involved in neuronal differentiation. J. Cell. Biochem 119, 3174–3182. - PubMed
-
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

