Machine learning demonstrates that somatic mutations imprint invariant morphologic features in myelodysplastic syndromes
- PMID: 32961553
- PMCID: PMC7702479
- DOI: 10.1182/blood.2020005488
Machine learning demonstrates that somatic mutations imprint invariant morphologic features in myelodysplastic syndromes
Abstract
Morphologic interpretation is the standard in diagnosing myelodysplastic syndrome (MDS), but it has limitations, such as varying reliability in pathologic evaluation and lack of integration with genetic data. Somatic events shape morphologic features, but the complexity of morphologic and genetic changes makes clear associations challenging. This article interrogates novel clinical subtypes of MDS using a machine-learning technique devised to identify patterns of cooccurrence among morphologic features and genomic events. We sequenced 1079 MDS patients and analyzed bone marrow morphologic alterations and other clinical features. A total of 1929 somatic mutations were identified. Five distinct morphologic profiles with unique clinical characteristics were defined. Seventy-seven percent of higher-risk patients clustered in profile 1. All lower-risk (LR) patients clustered into the remaining 4 profiles: profile 2 was characterized by pancytopenia, profile 3 by monocytosis, profile 4 by elevated megakaryocytes, and profile 5 by erythroid dysplasia. These profiles could also separate patients with different prognoses. LR MDS patients were classified into 8 genetic signatures (eg, signature A had TET2 mutations, signature B had both TET2 and SRSF2 mutations, and signature G had SF3B1 mutations), demonstrating association with specific morphologic profiles. Six morphologic profiles/genetic signature associations were confirmed in a separate analysis of an independent cohort. Our study demonstrates that nonrandom or even pathognomonic relationships between morphology and genotype to define clinical features can be identified. This is the first comprehensive implementation of machine-learning algorithms to elucidate potential intrinsic interdependencies among genetic lesions, morphologies, and clinical prognostic in attributes of MDS.
© 2020 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: B.P.H. reports research funds from Amgen and is a scientific advisor for Presagia. The remaining authors declare no competing financial interests.
Figures

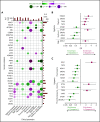
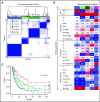
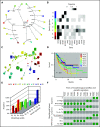
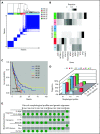
Comment in
-
Human and artificial intelligence to illuminate MDS.Blood. 2020 Nov 12;136(20):2243-2244. doi: 10.1182/blood.2020008742. Blood. 2020. PMID: 33180921 No abstract available.
References
-
- Vardiman JW, Thiele J, Arber DA, et al. . The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937-951. - PubMed
-
- Arber DA, Orazi A, Hasserjian R, et al. . The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia [published correction appears in Blood. 2016;128(3):462-463]. Blood. 2016;127(20):2391-2405. - PubMed
-
- Stephenson J, Mufti GJ, Yoshida Y. Myelodysplastic syndromes: from morphology to molecular biology. Part II. The molecular genetics of myelodysplasia. Int J Hematol. 1993;57(2):99-112. - PubMed
-
- Cazzola M, Della Porta MG, Travaglino E, Malcovati L. Classification and prognostic evaluation of myelodysplastic syndromes. Semin Oncol. 2011;38(5):627-634. - PubMed
-
- Greenberg P, Cox C, LeBeau MM, et al. . International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89(6):2079-2088. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

