Differential Gene Expression with an Emphasis on Floral Organ Size Differences in Natural and Synthetic Polyploids of Nicotiana tabacum (Solanaceae)
- PMID: 32961813
- PMCID: PMC7563459
- DOI: 10.3390/genes11091097
Differential Gene Expression with an Emphasis on Floral Organ Size Differences in Natural and Synthetic Polyploids of Nicotiana tabacum (Solanaceae)
Abstract
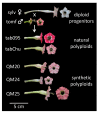
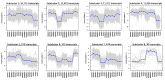
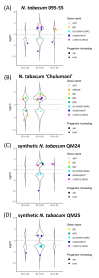
Floral organ size, especially the size of the corolla, plays an important role in plant reproduction by facilitating pollination efficiency. Previous studies have outlined a hypothesized organ size pathway. However, the expression and function of many of the genes in the pathway have only been investigated in model diploid species; therefore, it is unknown how these genes interact in polyploid species. Although correlations between ploidy and cell size have been shown in many systems, it is unclear whether there is a difference in cell size between naturally occurring and synthetic polyploids. To address these questions comparing floral organ size and cell size across ploidy, we use natural and synthetic polyploids of Nicotiana tabacum (Solanaceae) as well as their known diploid progenitors. We employ a comparative transcriptomics approach to perform analyses of differential gene expression, focusing on candidate genes that may be involved in floral organ size, both across developmental stages and across accessions. We see differential expression of several known floral organ candidate genes including ARF2, BIG BROTHER, and GASA/GAST1. Results from linear models show that ploidy, cell width, and cell number positively influence corolla tube circumference; however, the effect of cell width varies by ploidy, and diploids have a significantly steeper slope than both natural and synthetic polyploids. These results demonstrate that polyploids have wider cells and that polyploidy significantly increases corolla tube circumference.
Keywords: BIG BROTHER; GASA; Nanopore; RNA-Seq; cell size; flower size.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

