The Transcriptomic Response of the Murine Thyroid Gland to Iodide Overload and the Role of the Nrf2 Antioxidant System
- PMID: 32961913
- PMCID: PMC7555824
- DOI: 10.3390/antiox9090884
The Transcriptomic Response of the Murine Thyroid Gland to Iodide Overload and the Role of the Nrf2 Antioxidant System
Abstract
Background: Thyroid follicular cells have physiologically high levels of reactive oxygen species because oxidation of iodide is essential for the iodination of thyroglobulin (Tg) during thyroid hormone synthesis. Thyroid follicles (the functional units of the thyroid) also utilize incompletely understood autoregulatory mechanisms to defend against exposure to excess iodide. To date, no transcriptomic studies have investigated these phenomena in vivo. Nuclear erythroid factor 2 like 2 (Nrf2 or Nfe2l2) is a transcription factor that regulates the expression of numerous antioxidant and other cytoprotective genes. We showed previously that the Nrf2 pathway regulates the antioxidant defense of follicular cells, as well as Tg transcription and Tg iodination. We, thus, hypothesized that Nrf2 might be involved in the transcriptional response to iodide overload.
Methods: C57BL6/J wild-type (WT) or Nrf2 knockout (KO) male mice were administered regular water or water supplemented with 0.05% sodium iodide for seven days. RNA from their thyroids was prepared for next-generation RNA sequencing (RNA-Seq). Gene expression changes were assessed and pathway analyses were performed on the sets of differentially expressed genes.
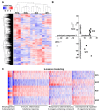
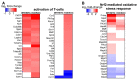
Results: Analysis of differentially expressed messenger RNAs (mRNAs) indicated that iodide overload upregulates inflammatory-, immune-, fibrosis- and oxidative stress-related pathways, including the Nrf2 pathway. Nrf2 KO mice showed a more pronounced inflammatory-autoimmune transcriptional response to iodide than WT mice. Compared to previously published datasets, the response patterns observed in WT mice had strong similarities with the patterns typical of Graves' disease and papillary thyroid carcinoma (PTC). Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) also responded to iodide overload, with the latter targeting mRNAs that participate mainly in inflammation pathways.
Conclusions: Iodide overload induces the Nrf2 cytoprotective response and upregulates inflammatory, immune, and fibrosis pathways similar to autoimmune hyperthyroidism (Graves' disease) and PTC.
Keywords: Kelch-like ECH-associated protein 1 (Keap1); Nfe2l2; RNA-Seq; fibrosis; immune response; inflammation; iodine; oxidative stress.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures








References
-
- Ziros P.G., Habeos I.G., Chartoumpekis D.V., Ntalampyra E., Somm E., Renaud C.O., Bongiovanni M., Trougakos I.P., Yamamoto M., Kensler T.W., et al. NFE2-Related Transcription Factor 2 Coordinates Antioxidant Defense with Thyroglobulin Production and Iodination in the Thyroid Gland. Thyroid. 2018;28:780–798. doi: 10.1089/thy.2018.0018. - DOI - PMC - PubMed
Grants and funding
- 31003A_153062, 31003A_182105, IZSEZ0_193623 and IZCOZ0_177070/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung
- 2016 Fellowship for Academic Promotion in Clinical Medicine/the Leenaards Foundation
- CA16112/European Cooperation in Science and Technology
- Exchange in Endocrinology Expertise (3E) Fellowship/Section/Board of Endocrinology of the European Union of Medical Specialists (UEMS)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

